|
Record Information |
|---|
| Version |
1.0 |
|---|
| Update Date |
1/22/2018 11:54:54 AM |
|---|
|
Metabolite ID | PAMDB120143 |
|---|
|
Identification |
|---|
| Name: |
2-(1,2-epoxy-1,2-dihydrophenyl)acetyl-CoA |
|---|
| Description: | An acyl-CoA(4−) oxoanion arising from deprotonation of the phosphate and diphosphate OH groups of 2-(1,2-epoxy-1,2-dihydrophenyl)acetyl-CoA; major species at pH 7.3. |
|---|
|
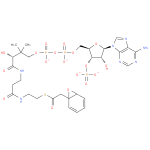
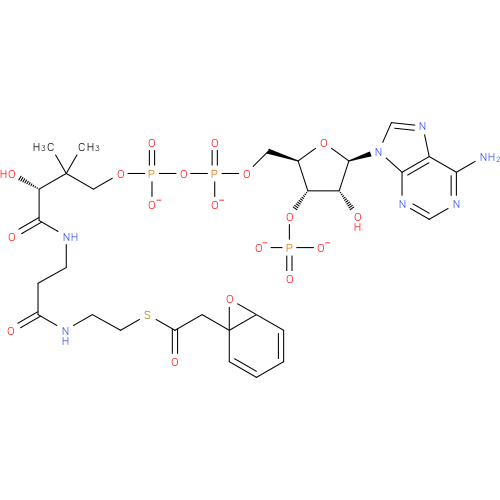
Structure |
|
|---|
| Synonyms: | - 2-(1,2-epoxy-1,2-dihydrophenyl)acetyl-CoA
- 2-{7-oxabicyclo[4.1.0]hepta-2,4-dien-1-yl}acetyl-CoA
|
|---|
|
Chemical Formula: |
C29H38N7O18P3S |
|---|
| Average Molecular Weight: |
897.637 |
|---|
| Monoisotopic Molecular
Weight: |
901.152 |
|---|
| InChI Key: |
ZTMHVINYLDVBNO-FOGVYBFTSA-J |
|---|
| InChI: | InChI=1S/C29H42N7O18P3S/c1-28(2,23(40)26(41)32-8-6-18(37)31-9-10-58-19(38)11-29-7-4-3-5-17(29)52-29)13-50-57(47,48)54-56(45,46)49-12-16-22(53-55(42,43)44)21(39)27(51-16)36-15-35-20-24(30)33-14-34-25(20)36/h3-5,7,14-17,21-23,27,39-40H,6,8-13H2,1-2H3,(H,31,37)(H,32,41)(H,45,46)(H,47,48)(H2,30,33,34)(H2,42,43,44)/p-4/t16-,17?,21-,22-,23+,27-,29?/m1/s1 |
|---|
| CAS
number: |
Not Available |
|---|
| IUPAC Name: | 3'- phosphonatoadenosine 5'- phosphonatoadenosine 5'- {3- {3- [(3R)- [(3R)- 4- 4- {[3- {[3- ({2- ({2- [(7- [(7- oxabicyclo[4.1.0]hepta- oxabicyclo[4.1.0]hepta- 2,4- 2,4- dien- dien- 1- 1- ylacetyl)sulfanyl]ethyl}amino)- ylacetyl)sulfanyl]ethyl}amino)- 3- 3- oxopropyl]amino}- oxopropyl]amino}- 3- 3- hydroxy- hydroxy- 2,2- 2,2- dimethyl- dimethyl- 4- 4- oxobutyl] diphosphate} oxobutyl] diphosphate} |
|---|
|
Traditional IUPAC Name: |
Not Available |
|---|
| SMILES: | CC(C)(C(O)C(=O)NCCC(=O)NCCSC(=O)CC12(OC1C=CC=C2))COP(=O)(OP(=O)(OCC3(C(OP([O-])(=O)[O-])C(O)C(O3)N5(C4(=C(C(N)=NC=N4)N=C5))))[O-])[O-] |
|---|
|
Chemical Taxonomy |
|---|
|
Taxonomy Description | This compound belongs to the class of organic compounds known as acyl coas. These are organic compounds containing a coenzyme A substructure linked to an acyl chain. |
|---|
|
Kingdom |
Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
|
Class |
Fatty Acyls |
|---|
| Sub Class | Fatty acyl thioesters |
|---|
|
Direct Parent |
Acyl CoAs |
|---|
| Alternative Parents |
|
|---|
| Substituents |
- Coenzyme a or derivatives
- Purine ribonucleoside 3',5'-bisphosphate
- Purine ribonucleoside bisphosphate
- Purine ribonucleoside diphosphate
- Ribonucleoside 3'-phosphate
- Pentose phosphate
- Pentose-5-phosphate
- Beta amino acid or derivatives
- Glycosyl compound
- N-glycosyl compound
- 6-aminopurine
- Organic pyrophosphate
- Monosaccharide phosphate
- Imidazopyrimidine
- Purine
- Aminopyrimidine
- N-substituted imidazole
- Organic phosphoric acid derivative
- N-acyl-amine
- Monosaccharide
- Phosphoric acid ester
- Fatty amide
- Imidolactam
- Alkyl phosphate
- Pyrimidine
- Azole
- Heteroaromatic compound
- Tetrahydrofuran
- Imidazole
- Carbothioic s-ester
- Carboxamide group
- Thiocarboxylic acid ester
- Amino acid or derivatives
- Secondary alcohol
- Secondary carboxylic acid amide
- Oxacycle
- Carboxylic acid derivative
- Azacycle
- Dialkyl ether
- Oxirane
- Ether
- Organoheterocyclic compound
- Sulfenyl compound
- Thiocarboxylic acid or derivatives
- Organopnictogen compound
- Organic oxygen compound
- Amine
- Alcohol
- Organic nitrogen compound
- Organic oxide
- Hydrocarbon derivative
- Carbonyl group
- Primary amine
- Organosulfur compound
- Organooxygen compound
- Organonitrogen compound
- Organic anion
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework |
Aromatic heteropolycyclic compounds |
|---|
| External Descriptors |
- a small molecule (CPD0-2362)
|
|---|
|
Physical Properties |
|---|
| State: |
Not Available |
|---|
| Charge: | -4 |
|---|
|
Melting point: |
Not Available |
|---|
| Experimental Properties: |
Not Available |
|---|
| Predicted Properties |
|
|---|
|
Biological Properties |
|---|
| Cellular Locations: |
Not Available |
|---|
| Reactions: | |
|---|
|
Pathways: |
- phenylacetate degradation I (aerobic)PWY0-321
|
|---|
|
Spectra |
|---|
| Spectra: |
Not Available |
|---|
|
References |
|---|
| References: |
- Ismail W, El-Said Mohamed M, Wanner BL, Datsenko KA, Eisenreich W, Rohdich F, Bacher A, Fuchs G (2003)Functional genomics by NMR spectroscopy. Phenylacetate catabolism in Escherichia coli. European journal of biochemistry 270, Pubmed: 12846838
- Teufel R, Mascaraque V, Ismail W, Voss M, Perera J, Eisenreich W, Haehnel W, Fuchs G (2010)Bacterial phenylalanine and phenylacetate catabolic pathway revealed. Proceedings of the National Academy of Sciences of the United States of America 107, Pubmed: 20660314
|
|---|
| Synthesis Reference: |
Not Available |
|---|
| Material Safety Data Sheet (MSDS) |
Not Available |
|---|
|
Links |
|---|
| External Links: |
|
|---|