|
Record Information |
|---|
| Version |
1.0 |
|---|
| Update Date |
1/22/2018 11:54:54 AM |
|---|
|
Metabolite ID | PAMDB120010 |
|---|
|
Identification |
|---|
| Name: |
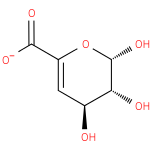
4-deoxy-β-L-threo-hex-4-enopyranuronose |
|---|
| Description: | A hexuronate that results from the removal of a proton from the carboxy group of 4-deoxy-β-L-threo-hex-4-enopyranuronic acid. |
|---|
|
Structure |
|
|---|
| Synonyms: | - (2S,3R,4S)-2,3,4-trihydroxy-3,4-dihydro-2H-pyran-6-carboxylate
- 4-deoxy-β-L-threo-hex-4-enopyranuronate
- 4-deoxy-β-L-threo-hex-4-enopyranuronate
- 4-deoxy-L-threo-5-hexosulose-uronate
|
|---|
|
Chemical Formula: |
C6H7O6 |
|---|
| Average Molecular Weight: |
175.118 |
|---|
| Monoisotopic Molecular
Weight: |
176.03209 |
|---|
| InChI Key: |
IAKKJSVSFCTLRY-YKKSOZKNSA-M |
|---|
| InChI: | InChI=1S/C6H8O6/c7-2-1-3(5(9)10)12-6(11)4(2)8/h1-2,4,6-8,11H,(H,9,10)/p-1/t2-,4+,6+/m0/s1 |
|---|
| CAS
number: |
Not Available |
|---|
| IUPAC Name: | 4-deoxy-β-L-threo-hex-4-enopyranuronate |
|---|
|
Traditional IUPAC Name: |
Not Available |
|---|
| SMILES: | C(C1(OC(C(C(C=1)O)O)O))([O-])=O |
|---|
|
Chemical Taxonomy |
|---|
|
Taxonomy Description | This compound belongs to the class of organic compounds known as hemiacetals. These are compounds comprising the hemiacetal functional group, with the general formula R2C(OH)OR' ( R' not Hydrogen ). |
|---|
|
Kingdom |
Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
|
Class |
Organooxygen compounds |
|---|
| Sub Class | Ethers |
|---|
|
Direct Parent |
Hemiacetals |
|---|
| Alternative Parents |
|
|---|
| Substituents |
- Secondary alcohol
- Hemiacetal
- 1,2-diol
- Oxacycle
- Organoheterocyclic compound
- Polyol
- Monocarboxylic acid or derivatives
- Carboxylic acid
- Carboxylic acid derivative
- Organic oxide
- Hydrocarbon derivative
- Carbonyl group
- Alcohol
- Organic anion
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework |
Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors |
- a modified sugar (CPD-13122)
|
|---|
|
Physical Properties |
|---|
| State: |
Not Available |
|---|
| Charge: | -1 |
|---|
|
Melting point: |
Not Available |
|---|
| Experimental Properties: |
Not Available |
|---|
| Predicted Properties |
|
|---|
|
Biological Properties |
|---|
| Cellular Locations: |
Not Available |
|---|
| Reactions: | |
|---|
|
Pathways: |
|
|---|
|
Spectra |
|---|
| Spectra: |
Not Available |
|---|
|
References |
|---|
| References: |
- Itoh T, Ochiai A, Mikami B, Hashimoto W, Murata K (2006)A novel glycoside hydrolase family 105: the structure of family 105 unsaturated rhamnogalacturonyl hydrolase complexed with a disaccharide in comparison with family 88 enzyme complexed with the disaccharide. Journal of molecular biology 360, Pubmed: 16781735
- Zhang R, Minh T, Lezondra L, Korolev S, Moy SF, Collart F, Joachimiak A (2005)1.6 A crystal structure of YteR protein from Bacillus subtilis, a predicted lyase. Proteins 60, Pubmed: 15906318
- Itoh T, Ochiai A, Mikami B, Hashimoto W, Murata K (2006)Structure of unsaturated rhamnogalacturonyl hydrolase complexed with substrate. Biochemical and biophysical research communications 347, Pubmed: 16870154
|
|---|
| Synthesis Reference: |
Not Available |
|---|
| Material Safety Data Sheet (MSDS) |
Not Available |
|---|
|
Links |
|---|
| External Links: |
|
|---|