|
Record Information |
|---|
| Version |
1.0 |
|---|
| Update Date |
1/22/2018 11:54:54 AM |
|---|
|
Metabolite ID | PAMDB110704 |
|---|
|
Identification |
|---|
| Name: |
FADH2 |
|---|
| Description: | The organophosphate oxoanion obtained by deprotonation of the diphosphate hydroxy groups of the reduced form of flavin adenine dinucleotide (FADH2). |
|---|
|
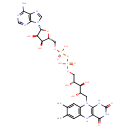
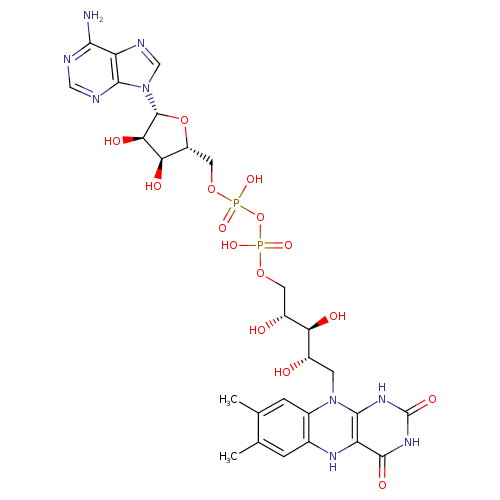
Structure |
|
|---|
| Synonyms: | -
flavin adenine dinucleotide reduced
-
1,5-dihydro-FAD
|
|---|
|
Chemical Formula: |
C27H33N9O15P2
|
|---|
| Average Molecular Weight: |
785.56 |
|---|
| Monoisotopic Molecular
Weight: |
787.1727845218 |
|---|
| InChI Key: |
YPZRHBJKEMOYQH-UYBVJOGSSA-L |
|---|
| InChI: |
InChI=1S/C27H35N9O15P2/c1-10-3-12-13(4-11(10)2)35(24-18(32-12)25(42)34-27(43)33-24)5-14(37)19(39)15(38)6-48-52(44,45)51-53(46,47)49-7-16-20(40)21(41)26(50-16)36-9-31-17-22(28)29-8-30-23(17)36/h3-4,8-9,14-16,19-21,26,32,37-41H,5-7H2,1-2H3,(H,44,45)(H,46,47)(H2,28,29,30)(H2,33,34,42,43)/p-2/t14-,15+,16+,19-,20+,21+,26+/m0/s1 |
|---|
| CAS
number: |
1910-41-4 |
|---|
| IUPAC Name: | adenosine 5'- (3- (3- {D- {D- ribo- ribo- 5- 5- [7,8- [7,8- dimethyl- dimethyl- 2,4- 2,4- dioxo- dioxo- 1,3,4,5- 1,3,4,5- tetrahydrobenzo[g]pteridin- tetrahydrobenzo[g]pteridin- 10(2H)- 10(2H)- yl]- yl]- 2,3,4- 2,3,4- trihydroxypentyl} diphosphate) trihydroxypentyl} diphosphate) |
|---|
|
Traditional IUPAC Name: |
fadh(.) |
|---|
| SMILES: | CC1(=C(C)C=C2(N(C3(NC(NC(=O)C(NC(=C1)2)=3)=O))CC(O)C(O)C(O)COP(OP([O-])(OCC6(C(O)C(O)C(N5(C=NC4(C(N)=NC=NC=45)))O6))=O)([O-])=O)) |
|---|
|
Chemical Taxonomy |
|---|
|
Taxonomy Description | This compound belongs to the class of chemical entities known as flavin nucleotides. These are nucleotides containing a flavin moiety. Flavin is a compound that contains the tricyclic isoalloxazine ring system, which bears 2 oxo groups at the 2- and 4-positions. |
|---|
|
Kingdom |
Chemical entities |
|---|
| Super Class | Organic compounds |
|---|
|
Class |
Nucleosides, nucleotides, and analogues |
|---|
| Sub Class | Flavin nucleotides |
|---|
|
Direct Parent |
Flavin nucleotides |
|---|
| Alternative Parents |
|
|---|
| Substituents |
- Flavin nucleotide
- Purine ribonucleoside diphosphate
- Purine ribonucleoside monophosphate
- Flavin
- Pentose-5-phosphate
- Pentose phosphate
- Alkyldiarylamine
- Glycosyl compound
- N-glycosyl compound
- 6-aminopurine
- Pteridine
- Pentose monosaccharide
- Monosaccharide phosphate
- Organic pyrophosphate
- Imidazopyrimidine
- Purine
- Monoalkyl phosphate
- Pyrimidone
- Aminopyrimidine
- Monosaccharide
- N-substituted imidazole
- Organic phosphoric acid derivative
- Imidolactam
- Benzenoid
- Phosphoric acid ester
- Primary aromatic amine
- Alkyl phosphate
- Pyrimidine
- Vinylogous amide
- Oxolane
- Azole
- Heteroaromatic compound
- Imidazole
- Secondary alcohol
- Urea
- Lactam
- Secondary amine
- Polyol
- Organoheterocyclic compound
- Azacycle
- Oxacycle
- Hydrocarbon derivative
- Organic nitrogen compound
- Alcohol
- Amine
- Organonitrogen compound
- Primary amine
- Organopnictogen compound
- Organic oxide
- Organooxygen compound
- Organic oxygen compound
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework |
Aromatic heteropolycyclic compounds |
|---|
| External Descriptors |
|
|---|
|
Physical Properties |
|---|
| State: |
Solid |
|---|
| Charge: | -2 |
|---|
|
Melting point: |
Not Available |
|---|
| Experimental Properties: |
| Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties |
|
|---|
|
Biological Properties |
|---|
| Cellular Locations: |
Not Available |
|---|
| Reactions: | |
|---|
|
Pathways: |
|
|---|
|
Spectra |
|---|
| Spectra: |
| Spectrum Type | Description | Splash Key | |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-000i-0001200900-b0740b3d33d50996ccde | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-000m-0105900000-3abff26d5bb35f8527b8 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-000i-0931700000-73f360589a13230eea7a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | Not Available |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | Not Available |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | Not Available |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | Not Available |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | Not Available |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | Not Available |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available |
|---|
| 2D NMR | [1H,1H] 2D NMR Spectrum | Not Available |
|---|
| 2D NMR | [1H,13C] 2D NMR Spectrum | Not Available |
|---|
|
|---|
|
References |
|---|
| References: |
- Zeller HD, Hille R, Jorns MS: Bacterial sarcosine oxidase: identification of novel substrates and a biradical reaction intermediate. Biochemistry. 1989 Jun 13;28(12):5145-54. [2475174 ]
- Ramsey AJ, Alderfer JL, Jorns MS: Energy transduction during catalysis by Escherichia coli DNA photolyase. Biochemistry. 1992 Aug 11;31(31):7134-42. [1643047 ]
- Ramsey AJ, Jorns MS: Effect of 5-deazaflavin on energy transduction during catalysis by Escherichia coli DNA photolyase. Biochemistry. 1992 Sep 15;31(36):8437-41. [1390627 ]
- Jorns MS: DNA photorepair: chromophore composition and function in two classes of DNA photolyases. Biofactors. 1990 Oct;2(4):207-11. [2282137 ]
|
|---|
| Synthesis Reference: |
Kavakli I Halil; Sancar Aziz Analysis of the role of intraprotein electron transfer in photoreactivation by DNA photolyase in vivo. Biochemistry (2004), 43(48), 15103-10. |
|---|
| Material Safety Data Sheet (MSDS) |
Not Available |
|---|
|
Links |
|---|
| External Links: |
|
|---|