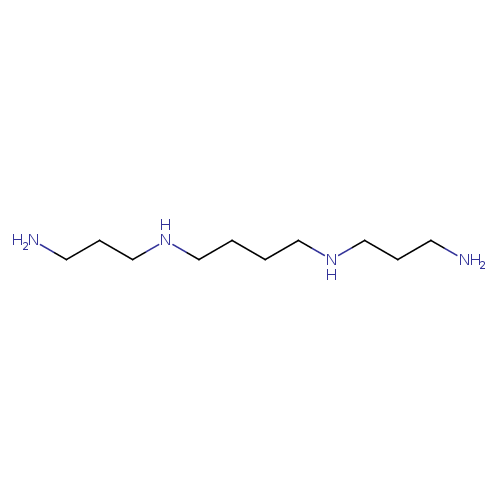
| Spectra: |
| Spectrum Type | Description | Splash Key | |
|---|
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Pegasus III TOF-MS system, Leco; GC 6890, Agilent Technologies) (x TMS) | splash10-014u-1900000000-e9620319823eaecceccc | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Pegasus III TOF-MS system, Leco; GC 6890, Agilent Technologies) (x TMS) | splash10-00s6-3900000000-d7da5cd1c0be289412ee | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Pegasus III TOF-MS system, Leco; GC 6890, Agilent Technologies) | splash10-00r6-1900000000-861e0541e2eb67219b93 | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Pegasus III TOF-MS system, Leco; GC 6890, Agilent Technologies) (5 TMS) | splash10-00di-7900000000-9706a16d903df3f9e094 | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Pegasus III TOF-MS system, Leco; GC 6890, Agilent Technologies) (5 TMS) | splash10-00di-5900000000-eb1574dc4aea2972932b | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - GC-MS (5 TMS) | splash10-00ei-8900000000-710d44c573ed398f7db9 | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - GC-MS (5 TMS) | splash10-00s6-3900000000-cb82c1371fa767015cff | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - GC-MS (6 TMS) | splash10-00rf-1900000000-ca3a2df44acb740134cd | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - GC-MS (6 TMS) | splash10-00y0-1900000000-7ba8e67861945901b381 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS | Not Available |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-0w29-2940000000-a8b05c7cfaea58c5d82d | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-03di-1900000000-75ac3953e5378c465a62 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-001i-9000000000-e3b84db5723912be8a4f | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-0udi-0930000000-4bb02118dea11e2101f8 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-0006-9000000000-468357ae06dea8985d85 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-00b9-6900000000-cf244bbfdbb5b7889d7a | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-004i-0900000000-c5af077deeab934c36b7 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-0udi-0940000000-098121f449eb29dd74be | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-0006-9000000000-167ef7503b0827eb212f | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-03di-0900000000-feb04e188e675948f795 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-004i-0900000000-93e15f4592fe9bbeb367 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 10V, Positive | splash10-0udi-0090000000-c22e98adb3dc62b1eb77 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 20V, Positive | splash10-01t9-0900000000-76aa5e4bf523c2027cba | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 30V, Positive | splash10-03di-4900000000-b762c20d8a09a78cf931 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 40V, Positive | splash10-001i-9300000000-0428cb6df2b593ded567 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 50V, Positive | splash10-001i-9000000000-c319f927047d78e2b69b | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-IT (LC/MSD Trap XCT, Agilent Technologies) , Positive | splash10-004i-0900000000-b4f106f6fe26766fbf73 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-IT (LC/MSD Trap XCT, Agilent Technologies) , Positive | splash10-03di-0900000000-29d4a223bb44ded927b5 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-IT (LC/MSD Trap XCT, Agilent Technologies) , Positive | splash10-01q9-9500000000-d1b7e914e9ccee2d0f66 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0udr-2970000000-268b93360fb5eaa19e49 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0a73-9810000000-ee919f22493191f56708 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a4l-9200000000-f7cb18a7a6036554cb6b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0udi-0090000000-ea2e2f233958fa61cd8b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0udi-3390000000-0ade7d389047531a84ee | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-05fu-9200000000-e760ec8b3259e54992ce | View in MoNA |
|---|
| MS | Mass Spectrum (Electron Ionization) | splash10-053u-9100000000-5a106273203d28e4a701 | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available |
|---|
| 2D NMR | [1H,1H] 2D NMR Spectrum | Not Available |
|---|
| 2D NMR | [1H,13C] 2D NMR Spectrum | Not Available |
|---|
|
|---|
| References: |
- Sreekumar A, Poisson LM, Rajendiran TM, Khan AP, Cao Q, Yu J, Laxman B, Mehra R, Lonigro RJ, Li Y, Nyati MK, Ahsan A, Kalyana-Sundaram S, Han B, Cao X, Byun J, Omenn GS, Ghosh D, Pennathur S, Alexander DC, Berger A, Shuster JR, Wei JT, Varambally S, Beecher C, Chinnaiyan AM: Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 2009 Feb 12;457(7231):910-4. [19212411 ]
- Mollica F, Li Volti S, Rapisarda A, Longo G, Pavone L, Vanella A: Increased erythrocytic spermine in Duchenne muscular dystrophy. Pediatr Res. 1980 Nov;14(11):1196-8. [7454431 ]
- Swift TA, Dias JA: Effects of the polyamine spermine on binding of follicle-stimulating hormone to membrane-bound immature bovine testis receptors. Biochim Biophys Acta. 1986 Feb 21;885(2):221-30. [3004602 ]
- Cipolla BG, Ziade J, Bansard JY, Moulinoux JP, Staerman F, Quemener V, Lobel B, Guille F: Pretherapeutic erythrocyte polyamine spermine levels discriminate high risk relapsing patients with M1 prostate carcinoma. Cancer. 1996 Sep 1;78(5):1055-65. [8780544 ]
- Venza M, Visalli M, Cicciu D, Teti D: Determination of polyamines in human saliva by high-performance liquid chromatography with fluorescence detection. J Chromatogr B Biomed Sci Appl. 2001 Jun 5;757(1):111-7. [11419735 ]
- Uehara N, Shirakawa S, Uchino H, Saeki Y: Elevated contents of spermidine and spermine in the erythrocytes of cancer patients. Cancer. 1980 Jan 1;45(1):108-11. [7350997 ]
- Jensen PK, Therkelsen AJ: Selective inhibition of fibroblasts by spermine in primary cultures of normal human skin epithelial cells. In Vitro. 1982 Oct;18(10):867-71. [7173947 ]
- Loser C, Folsch UR, Paprotny C, Creutzfeldt W: Polyamines in colorectal cancer. Evaluation of polyamine concentrations in the colon tissue, serum, and urine of 50 patients with colorectal cancer. Cancer. 1990 Feb 15;65(4):958-66. [2297664 ]
- Proctor MS, Fletcher HV Jr, Shukla JB, Rennert OM: Elevated spermidine and spermine levels in the blood of psoriasis patients. J Invest Dermatol. 1975 Oct;65(4):409-11. [1176793 ]
- Lorenz B, Francis F, Gempel K, Boddrich A, Josten M, Schmahl W, Schmidt J, Lehrach H, Meitinger T, Strom TM: Spermine deficiency in Gy mice caused by deletion of the spermine synthase gene. Hum Mol Genet. 1998 Mar;7(3):541-7. [9467015 ]
- Hosseinkhani H, Azzam T, Tabata Y, Domb AJ: Dextran-spermine polycation: an efficient nonviral vector for in vitro and in vivo gene transfection. Gene Ther. 2004 Jan;11(2):194-203. [14712304 ]
- El Baze P, Milano G, Verrando P, Renee N, Ortonne JP: Polyamine levels in normal human skin. A comparative study of pure epidermis, pure dermis, and suction blister fluid. Arch Dermatol Res. 1983;275(4):218-21. [6625645 ]
|
|---|