|
Record Information |
|---|
| Version |
1.0 |
|---|
| Update Date |
1/22/2018 11:54:54 AM |
|---|
|
Metabolite ID | PAMDB110497 |
|---|
|
Identification |
|---|
| Name: |
5,6-dihydrouracil |
|---|
| Description: | A pyrimidine obtained by formal addition of hydrogen across the 5,6-position of uracil. |
|---|
|
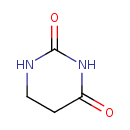
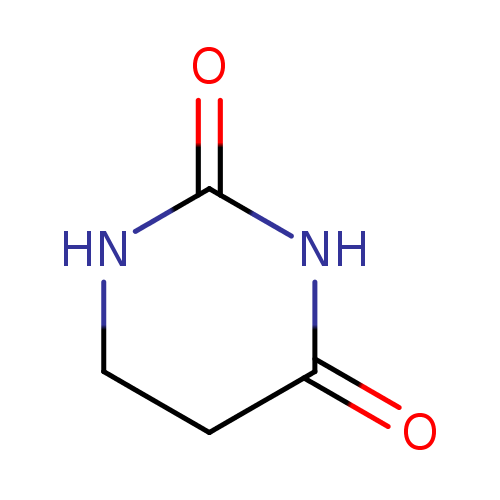
Structure |
|
|---|
| Synonyms: | -
dihydrouracil
-
DI-H-uracil
|
|---|
|
Chemical Formula: |
C4H6N2O2
|
|---|
| Average Molecular Weight: |
114.1 |
|---|
| Monoisotopic Molecular
Weight: |
114.0429274472 |
|---|
| InChI Key: |
OIVLITBTBDPEFK-UHFFFAOYSA-N |
|---|
| InChI: |
InChI=1S/C4H6N2O2/c7-3-1-2-5-4(8)6-3/h1-2H2,(H2,5,6,7,8) |
|---|
| CAS
number: |
504-07-4 |
|---|
| IUPAC Name: | dihydropyrimidine-2,4(1H,3H)-dione |
|---|
|
Traditional IUPAC Name: |
dihydrouracil |
|---|
| SMILES: | C1(NC(CCN1)=O)=O |
|---|
|
Chemical Taxonomy |
|---|
|
Taxonomy Description | This compound belongs to the class of chemical entities known as pyrimidones. These are compounds that contain a pyrimidine ring, which bears a ketone. Pyrimidine is a 6-membered ring consisting of four carbon atoms and two nitrogen centers at the 1- and 3- ring positions. |
|---|
|
Kingdom |
Chemical entities |
|---|
| Super Class | Organic compounds |
|---|
|
Class |
Organoheterocyclic compounds |
|---|
| Sub Class | Diazines |
|---|
|
Direct Parent |
Pyrimidones |
|---|
| Alternative Parents |
|
|---|
| Substituents |
- Pyrimidone
- Ureide
- 1,3-diazinane
- Dicarboximide
- Urea
- Azacycle
- Carboxylic acid derivative
- Organic oxide
- Organopnictogen compound
- Organooxygen compound
- Organonitrogen compound
- Organic oxygen compound
- Organic nitrogen compound
- Carbonyl group
- Hydrocarbon derivative
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework |
Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors |
|
|---|
|
Physical Properties |
|---|
| State: |
Solid |
|---|
| Charge: | 0 |
|---|
|
Melting point: |
279 - 281 °C |
|---|
| Experimental Properties: |
| Property | Value | Reference |
|---|
| Melting Point | 279 - 281 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties |
|
|---|
|
Biological Properties |
|---|
| Cellular Locations: |
Not Available |
|---|
| Reactions: | |
|---|
|
Pathways: |
|
|---|
|
Spectra |
|---|
| Spectra: |
| Spectrum Type | Description | Splash Key | |
|---|
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Pegasus III TOF-MS system, Leco; GC 6890, Agilent Technologies) (2 TMS) | splash10-00di-9700000000-4adeeb2f4f5a1111afb7 | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - GC-MS (1 TMS) | splash10-00di-5900000000-8af1b81502646fa5b417 | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - GC-MS (2 TMS) | splash10-0fdo-9750000000-d2aaeeba1cf962175f84 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-03k9-9800000000-da772cea603c785c1df4 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-0a4i-9100000000-94a5f543e8c6be7b3204 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-0btc-9200000000-e9bf2ae82714a2a3a737 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Positive | splash10-014i-5900000000-0fe83e63f6e5d1347894 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-014i-2900000000-ad7f91d5265e53403dd6 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0603-9200000000-4b30a83fb454e5bbffb8 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0006-9000000000-ce54e3c860a95b0af401 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-03di-9800000000-d705e188f6700a76ef72 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0006-9000000000-a26409d69a4259731004 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9000000000-f00c4df547660023bee4 | View in MoNA |
|---|
| MS | Mass Spectrum (Electron Ionization) | splash10-03fu-9400000000-5b9edda3479e7d0a0d28 | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available |
|---|
| 2D NMR | [1H,13C] 2D NMR Spectrum | Not Available |
|---|
|
|---|
|
References |
|---|
| References: |
- Roux A, Xu Y, Heilier JF, Olivier MF, Ezan E, Tabet JC, Junot C (2012)Annotation of the human adult urinary metabolome and metabolite identification using ultra high performance liquid chromatography coupled to a linear quadrupole ion trap-Orbitrap mass spectrometer. Analytical chemistry 84, Pubmed: 22770225
- Aoun R, Renaud JL, Dixneuf PH, Bruneau C (2005)Concomitant monoreduction and hydrogenation of unsaturated cyclic imides to lactams catalyzed by ruthenium compounds. Angewandte Chemie (International ed. in English) 44, Pubmed: 15724255
- Kristensen MH, Pedersen P, Mejer J (2010)The value of dihydrouracil/uracil plasma ratios in predicting 5-fluorouracil-related toxicity in colorectal cancer patients. The Journal of international medical research 38, Pubmed: 20926004
- Ben Fredj R, Gross E, Ben Ahmed S, Hassine H, Saguem S (2009)The dihydrouracil/uracil ratio in plasma, clinical and genetic analysis for screening of dihydropyrimidine dehydrogenase deficiency in colorectal cancer patients treated with 5-fluorouracil. Pathologie-biologie 57, Pubmed: 18619742
|
|---|
| Synthesis Reference: |
Bhat, K. S.; Rao, A. S. Synthesis of uracil, 6-methyluracil and some dihydrouracils. Organic Preparations and Procedures International (1983), 15(5), 303-13. |
|---|
| Material Safety Data Sheet (MSDS) |
Download (PDF) |
|---|
|
Links |
|---|
| External Links: |
|
|---|