Record Information Version
1.0 Update Date
1/22/2018 11:54:54 AM
Metabolite ID PAMDB110492
Identification Name:
queuine Description: Not Available
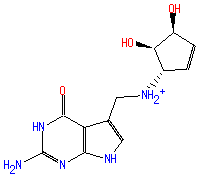
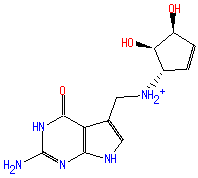
Structure
Synonyms:
7-(3,4-trans-4,5-cis-dihydroxy-1-cyclopenten-3-ylaminomethyl)-7-deazaguanine
base Q
4H-pyrrolo(2,3-d)pyrimidin-4-one 2-amino-5-((((1S,4S,5R)-4,5-dihydroxy-2-cyclopenten-1-yl)amino)methyl)-1,7-dihydro-
Chemical Formula:
C12 H16 N5 O3
Average Molecular Weight:
278.1253144059 Monoisotopic Molecular
Weight:
278.1253144059 InChI Key:
WYROLENTHWJFLR-ACLDMZEESA-O InChI:
InChI=1S/C12H15N5O3/c13-12-16-10-8(11(20)17-12)5(4-15-10)3-14-6-1-2-7(18)9(6)19/h1-2,4,6-7,9,14,18-19H,3H2,(H4,13,15,16,17,20)/p+1/t6-,7-,9+/m0/s1 CAS
number:
Not Available IUPAC Name: Not Available
Traditional IUPAC Name:
Not Available SMILES: C([N+]C1(C=CC(O)C(O)1))C2(=CNC3(N=C(NC(=O)C2=3)N))
Chemical Taxonomy
Taxonomy Description This compound belongs to the class of organic compounds known as pyrrolo[2,3-d]pyrimidines. These are aromatic heteropolycyclic compounds containing a pyrrolo[2,3-d]pyrimidine ring system, which is an pyrrolopyrimidine isomers having the 3 ring nitrogen atoms at the 1-, 5-, and 7-positions.
Kingdom
Organic compounds Super Class Organoheterocyclic compounds
Class
Pyrrolopyrimidines Sub Class Pyrrolo[2,3-d]pyrimidines
Direct Parent
Pyrrolo[2,3-d]pyrimidines Alternative Parents
Substituents
Not Available Molecular Framework
Aromatic heteropolycyclic compounds External Descriptors
Not Available
Physical Properties State:
Not Available Charge: Not Available
Melting point:
Not Available Experimental Properties:
Not Available Predicted Properties
Biological Properties Cellular Locations:
Not Available Reactions:
Pathways:
Spectra Spectra:
Not Available
References References:
Not Available Synthesis Reference:
Not Available Material Safety Data Sheet (MSDS)
Not Available
Links External Links:
This project is supported by
the University of Maryland ,
School of Pharmacy ,
Mass Spectrometry Center , a Waters Center of Excellence. The center is an NIH-investigator funded research and core facility that supports a wide range of cutting-edge metabolomic studies. The Center is supported through a center grant from the University of Maryland and NIH grants to its members. The PAMDB project is affiliated with
The Metabolomics Innovation Centre (TMIC) a leading metabolomics research and service center funded through Genome Alberta, Genome British Columbia and Genome Canada.