|
Record Information |
|---|
| Version |
1.0 |
|---|
| Update Date |
1/22/2018 11:54:54 AM |
|---|
|
Metabolite ID | PAMDB110485 |
|---|
|
Identification |
|---|
| Name: |
XMP |
|---|
| Description: | A nucleoside 5'-monophosphate(2−) that results from the removal of two protons from the phosphate group of 5'-xanthylic acid; major species at pH 7.3. |
|---|
|
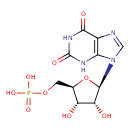
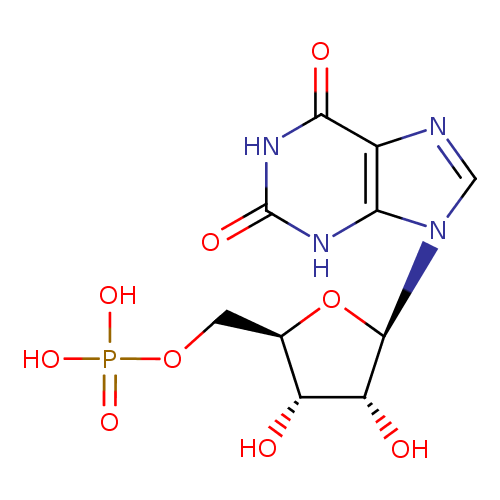
Structure |
|
|---|
| Synonyms: | -
(9-D-ribosylxanthine) 5'-phosphate
-
xanthosine 5-phosphate
-
xanthosine 5'-phosphate
-
xanthosine-5-P
-
xanthosine-5'-P
-
9-(5-phospho-β-D-ribosyl)xanthine
|
|---|
|
Chemical Formula: |
C10H11N4O9P
|
|---|
| Average Molecular Weight: |
362.19 |
|---|
| Monoisotopic Molecular
Weight: |
364.042014547 |
|---|
| InChI Key: |
DCTLYFZHFGENCW-UUOKFMHZSA-L |
|---|
| InChI: |
InChI=1S/C10H13N4O9P/c15-5-3(1-22-24(19,20)21)23-9(6(5)16)14-2-11-4-7(14)12-10(18)13-8(4)17/h2-3,5-6,9,15-16H,1H2,(H2,19,20,21)(H2,12,13,17,18)/p-2/t3-,5-,6-,9-/m1/s1 |
|---|
| CAS
number: |
523-98-8 |
|---|
| IUPAC Name: | 5'-O-phosphonatoxanthosine |
|---|
|
Traditional IUPAC Name: |
xanthosine monophosphate |
|---|
| SMILES: | C(OP(=O)([O-])[O-])C1(C(O)C(O)C(O1)N3(C=NC2(C(=O)NC(=O)NC=23))) |
|---|
|
Chemical Taxonomy |
|---|
|
Taxonomy Description | This compound belongs to the class of chemical entities known as purine ribonucleoside monophosphates. These are nucleotides consisting of a purine base linked to a ribose to which one monophosphate group is attached. |
|---|
|
Kingdom |
Chemical entities |
|---|
| Super Class | Organic compounds |
|---|
|
Class |
Nucleosides, nucleotides, and analogues |
|---|
| Sub Class | Purine nucleotides |
|---|
|
Direct Parent |
Purine ribonucleoside monophosphates |
|---|
| Alternative Parents |
|
|---|
| Substituents |
- Purine ribonucleoside monophosphate
- Pentose phosphate
- Pentose-5-phosphate
- Glycosyl compound
- N-glycosyl compound
- Xanthine
- 6-oxopurine
- Monosaccharide phosphate
- Pentose monosaccharide
- Purinone
- Alkaloid or derivatives
- Imidazopyrimidine
- Purine
- Monoalkyl phosphate
- Pyrimidone
- Alkyl phosphate
- Monosaccharide
- N-substituted imidazole
- Organic phosphoric acid derivative
- Phosphoric acid ester
- Pyrimidine
- Azole
- Heteroaromatic compound
- Oxolane
- Imidazole
- Vinylogous amide
- Secondary alcohol
- Urea
- 1,2-diol
- Lactam
- Oxacycle
- Organoheterocyclic compound
- Azacycle
- Alcohol
- Organonitrogen compound
- Organooxygen compound
- Organic oxygen compound
- Hydrocarbon derivative
- Organopnictogen compound
- Organic nitrogen compound
- Organic oxide
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework |
Aromatic heteropolycyclic compounds |
|---|
| External Descriptors |
|
|---|
|
Physical Properties |
|---|
| State: |
Solid |
|---|
| Charge: | -2 |
|---|
|
Melting point: |
Not Available |
|---|
| Experimental Properties: |
| Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties |
|
|---|
|
Biological Properties |
|---|
| Cellular Locations: |
Not Available |
|---|
| Reactions: | |
|---|
|
Pathways: |
|
|---|
|
Spectra |
|---|
| Spectra: |
|
|---|
|
References |
|---|
| References: |
- Wolan DW, Cheong CG, Greasley SE, Wilson IA: Structural insights into the human and avian IMP cyclohydrolase mechanism via crystal structures with the bound XMP inhibitor. Biochemistry. 2004 Feb 10;43(5):1171-83. [14756553 ]
- Vethe NT, Bergan S: Determination of inosine monophosphate dehydrogenase activity in human CD4+ cells isolated from whole blood during mycophenolic acid therapy. Ther Drug Monit. 2006 Oct;28(5):608-13. [17038874 ]
- Khalil PN, Erb N, Khalil MN, Escherich G, Janka-Schaub GE: Validation and application of a high-performance liquid chromatographic-based assay for determination of the inosine 5'-monophosphate dehydrogenase activity in erythrocytes. J Chromatogr B Analyt Technol Biomed Life Sci. 2006 Sep 14;842(1):1-7. Epub 2006 May 24. [16725387 ]
- Barsotti C, Pesi R, Giannecchini M, Ipata PL: Evidence for the involvement of cytosolic 5'-nucleotidase (cN-II) in the synthesis of guanine nucleotides from xanthosine. J Biol Chem. 2005 Apr 8;280(14):13465-9. Epub 2005 Feb 6. [15699053 ]
- Prosise GL, Luecke H: Crystal structures of Tritrichomonasfoetus inosine monophosphate dehydrogenase in complex with substrate, cofactor and analogs: a structural basis for the random-in ordered-out kinetic mechanism. J Mol Biol. 2003 Feb 14;326(2):517-27. [12559919 ]
- Stoychev G, Kierdaszuk B, Shugar D: Xanthosine and xanthine. Substrate properties with purine nucleoside phosphorylases, and relevance to other enzyme systems. Eur J Biochem. 2002 Aug;269(16):4048-57. [12180982 ]
- Daxecker H, Raab M, Muller MM: Influence of mycophenolic acid on inosine 5'-monophosphate dehydrogenase activity in human peripheral blood mononuclear cells. Clin Chim Acta. 2002 Apr;318(1-2):71-7. [11880114 ]
- Rauniyar RK, Suzuma K, King AL, Aiello LP, King GL: Differential effects of bactericidal/permeability-increasing protein (BPI) analogues on retinal neovascularization and retinal pericyte growth. Invest Ophthalmol Vis Sci. 2002 Feb;43(2):503-9. [11818397 ]
- Glander P, Braun KP, Hambach P, Bauer S, Mai I, Roots I, Waiser J, Fritsche L, Neumayer HH, Budde K: Non-radioactive determination of inosine 5'-monophosphate dehydro-genase (IMPDH) in peripheral mononuclear cells. Clin Biochem. 2001 Oct;34(7):543-9. [11738390 ]
- Frueh FW, Hayashibara KC, Brown PO, Whitlock JP Jr: Use of cDNA microarrays to analyze dioxin-induced changes in human liver gene expression. Toxicol Lett. 2001 Jul 6;122(3):189-203. [11489354 ]
- Wall M, Shim JH, Benkovic SJ: Human AICAR transformylase: role of the 4-carboxamide of AICAR in binding and catalysis. Biochemistry. 2000 Sep 19;39(37):11303-11. [10985775 ]
- Sintchak MD, Nimmesgern E: The structure of inosine 5'-monophosphate dehydrogenase and the design of novel inhibitors. Immunopharmacology. 2000 May;47(2-3):163-84. [10878288 ]
- Albrecht W, Storck M, Pfetsch E, Martin W, Abendroth D: Development and application of a high-performance liquid chromatography-based assay for determination of the activity of inosine 5'-monophosphate dehydrogenase in whole blood and isolated mononuclear cells. Ther Drug Monit. 2000 Jun;22(3):283-94. [10850395 ]
- McMillan FM, Cahoon M, White A, Hedstrom L, Petsko GA, Ringe D: Crystal structure at 2.4 A resolution of Borrelia burgdorferi inosine 5'-monophosphate dehydrogenase: evidence of a substrate-induced hinged-lid motion by loop 6. Biochemistry. 2000 Apr 18;39(15):4533-42. [10758003 ]
- Markland W, McQuaid TJ, Jain J, Kwong AD: Broad-spectrum antiviral activity of the IMP dehydrogenase inhibitor VX-497: a comparison with ribavirin and demonstration of antiviral additivity with alpha interferon. Antimicrob Agents Chemother. 2000 Apr;44(4):859-66. [10722482 ]
- Digits JA, Hedstrom L: Drug selectivity is determined by coupling across the NAD+ site of IMP dehydrogenase. Biochemistry. 2000 Feb 22;39(7):1771-7. [10677226 ]
- Digits JA, Hedstrom L: Species-specific inhibition of inosine 5'-monophosphate dehydrogenase by mycophenolic acid. Biochemistry. 1999 Nov 16;38(46):15388-97. [10563825 ]
- Heroux A, White EL, Ross LJ, Davis RL, Borhani DW: Crystal structure of Toxoplasma gondii hypoxanthine-guanine phosphoribosyltransferase with XMP, pyrophosphate, and two Mg(2+) ions bound: insights into the catalytic mechanism. Biochemistry. 1999 Nov 2;38(44):14495-506. [10545171 ]
- Heroux A, White EL, Ross LJ, Borhani DW: Crystal structures of the Toxoplasma gondii hypoxanthine-guanine phosphoribosyltransferase-GMP and -IMP complexes: comparison of purine binding interactions with the XMP complex. Biochemistry. 1999 Nov 2;38(44):14485-94. [10545170 ]
- Pitera JW, Munagala NR, Wang CC, Kollman PA: Understanding substrate specificity in human and parasite phosphoribosyltransferases through calculation and experiment. Biochemistry. 1999 Aug 10;38(32):10298-306. [10441123 ]
- Minakawa N, Matsuda A: Mechanism-based design of inosine 5-monophosphate dehydrogenase inhibitors: synthesis and biological activities of 5-ethynyl-1-beta-D-ribofuranosylimidazole-4-carboxamide (EICAR) and its derivatives. Curr Med Chem. 1999 Jul;6(7):615-28. [10390604 ]
- Franchetti P, Grifantini M: Nucleoside and non-nucleoside IMP dehydrogenase inhibitors as antitumor and antiviral agents. Curr Med Chem. 1999 Jul;6(7):599-614. [10390603 ]
- Jayaram HN, Cooney DA, Grusch M, Krupitza G: Consequences of IMP dehydrogenase inhibition, and its relationship to cancer and apoptosis. Curr Med Chem. 1999 Jul;6(7):561-74. [10390601 ]
- Hedstrom L: IMP dehydrogenase: mechanism of action and inhibition. Curr Med Chem. 1999 Jul;6(7):545-60. [10390600 ]
|
|---|
| Synthesis Reference: |
Hattori, Kyoji; Kawahara, Shin; Hagiwara, Takeshige. 5'-Xanthylic acid. Jpn. Kokai Tokkyo Koho (1985), 3 pp. |
|---|
| Material Safety Data Sheet (MSDS) |
Not Available |
|---|
|
Links |
|---|
| External Links: |
|
|---|