|
Record Information |
|---|
| Version |
1.0 |
|---|
| Update Date |
1/22/2018 11:54:54 AM |
|---|
|
Metabolite ID | PAMDB110409 |
|---|
|
Identification |
|---|
| Name: |
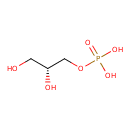
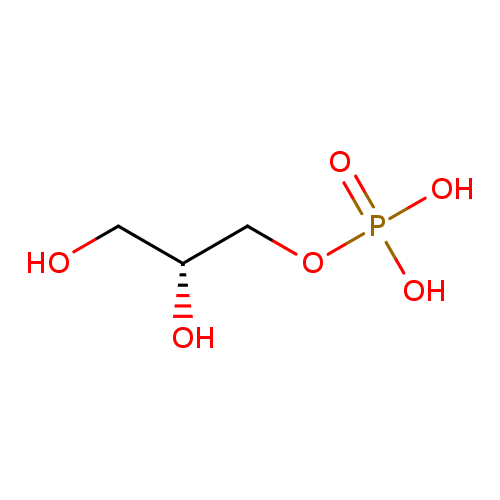
sn-glycerol 3-phosphate |
|---|
| Description: | An organophosphate oxoanion that is the dianion of sn-glycerol 3-phosphate arising from deprotonation of both phosphate OH groups. |
|---|
|
Structure |
|
|---|
| Synonyms: | -
L-α-glycerophosphate
-
L-G3P
-
L-glycerol 3-phosphate
-
α-glycerophosphoric acid
-
α-glycerophosphate
-
D-glycerol 1-phosphate
-
glycerol 3-phosphate
|
|---|
|
Chemical Formula: |
C3H7O6P
|
|---|
| Average Molecular Weight: |
170.06 |
|---|
| Monoisotopic Molecular
Weight: |
172.0136745315 |
|---|
| InChI Key: |
AWUCVROLDVIAJX-GSVOUGTGSA-L |
|---|
| InChI: |
InChI=1S/C3H9O6P/c4-1-3(5)2-9-10(6,7)8/h3-5H,1-2H2,(H2,6,7,8)/p-2/t3-/m1/s1 |
|---|
| CAS
number: |
57-03-4 |
|---|
| IUPAC Name: | (2R)-2,3-dihydroxypropyl phosphate |
|---|
|
Traditional IUPAC Name: |
3-phosphoglycerol |
|---|
| SMILES: | C(OP([O-])(=O)[O-])C(O)CO |
|---|
|
Chemical Taxonomy |
|---|
|
Taxonomy Description | This compound belongs to the class of chemical entities known as glycerophosphates. These are compounds containing a glycerol linked to a phosphate group. |
|---|
|
Kingdom |
Chemical entities |
|---|
| Super Class | Organic compounds |
|---|
|
Class |
Lipids and lipid-like molecules |
|---|
| Sub Class | Glycerophospholipids |
|---|
|
Direct Parent |
Glycerophosphates |
|---|
| Alternative Parents |
|
|---|
| Substituents |
- Sn-glycerol-3-phosphate
- Monoalkyl phosphate
- Alkyl phosphate
- Phosphoric acid ester
- Organic phosphoric acid derivative
- Secondary alcohol
- 1,2-diol
- Organic oxygen compound
- Organic oxide
- Hydrocarbon derivative
- Primary alcohol
- Organooxygen compound
- Alcohol
- Aliphatic acyclic compound
|
|---|
| Molecular Framework |
Aliphatic acyclic compounds |
|---|
| External Descriptors |
|
|---|
|
Physical Properties |
|---|
| State: |
Solid |
|---|
| Charge: | -2 |
|---|
|
Melting point: |
102 - 104 °C |
|---|
| Experimental Properties: |
| Property | Value | Reference |
|---|
| Melting Point | 102 - 104 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | 1000.0 mg/mL | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties |
|
|---|
|
Biological Properties |
|---|
| Cellular Locations: |
Not Available |
|---|
| Reactions: | |
|---|
|
Pathways: |
|
|---|
|
Spectra |
|---|
| Spectra: |
| Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS | Not Available |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-0002-9200000000-bf06c1d1e56cb8853243 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-00kr-9200000000-ffce45222cab00a0c302 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-001j-9000000000-7689cca8f9c9a12d9e75 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-00di-0902000000-00e3a7e1ee10fbd8b03e | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-000i-9000000000-1a40c90fd2eb656ddf5c | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-0002-9000000000-f9bc5f552db38767c437 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-0002-0920000000-72f8b96355726dd3a5b1 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-00di-0901000000-2c3a8ed438d7ca0fa049 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-000i-9000000000-a3549fb5ec2675fbf615 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-0002-9000000000-096e2ba9a6ef6fd20d88 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-0002-0930000000-171708905bad289d6031 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-00di-0925110000-c1ced10d34f533b3fa1f | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-03di-0900000000-4ee8a36492a451beda75 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-004i-9000000000-de2aacfb81476c7755e5 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-00di-0900000000-dd342482ade09e9ce749 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-0fkc-0917520000-d61e6034525ac5f7588d | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-004i-9000000000-d537d3f23f3d9a789c20 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-00di-0900000000-2f1ca45b50402f6e62df | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-0udi-0009000000-5606781224d6ab5caed3 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 10V, Positive | splash10-0pi0-1900000000-75b6ed57a4c9cd6378b4 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 20V, Positive | splash10-0a59-6900000000-1c46fbda1468f6fd803c | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 30V, Positive | splash10-0a5c-9400000000-305e9a2554d8c3274f32 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 40V, Positive | splash10-007o-9000000000-d11ad04a048c3dca25de | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 50V, Positive | splash10-00mo-9000000000-7b779b4be184b5b26b6a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | Not Available |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available |
|---|
| 2D NMR | [1H,1H] 2D NMR Spectrum | Not Available |
|---|
| 2D NMR | [1H,13C] 2D NMR Spectrum | Not Available |
|---|
|
|---|
|
References |
|---|
| References: |
- Sreekumar A, Poisson LM, Rajendiran TM, Khan AP, Cao Q, Yu J, Laxman B, Mehra R, Lonigro RJ, Li Y, Nyati MK, Ahsan A, Kalyana-Sundaram S, Han B, Cao X, Byun J, Omenn GS, Ghosh D, Pennathur S, Alexander DC, Berger A, Shuster JR, Wei JT, Varambally S, Beecher C, Chinnaiyan AM: Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 2009 Feb 12;457(7231):910-4. [19212411 ]
- Slomiany BL, Piotrowski J, Mojtahed H, Slomiany A: Ebrotidine effect on the proteolytic and lipolytic activities of Helicobacter pylori. Gen Pharmacol. 1992 Mar;23(2):203-6. [1639232 ]
- Guminski T: Some glycolytic enzymes in normal cerebrospinal fluid, brain tissue and blood plasma of infants. Clin Chim Acta. 1976 Aug 16;71(1):61-6. [134854 ]
|
|---|
| Synthesis Reference: |
Rios-Mercadillo, Victor M.; Whitesides, George M. Enzymic synthesis of sn-glycerol 3-phosphate. Journal of the American Chemical Society (1979), 101(19), 5828-9. |
|---|
| Material Safety Data Sheet (MSDS) |
Not Available |
|---|
|
Links |
|---|
| External Links: |
|
|---|