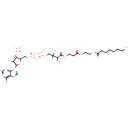|
Record Information |
|---|
| Version |
1.0 |
|---|
| Update Date |
1/22/2018 11:54:54 AM |
|---|
|
Metabolite ID | PAMDB110393 |
|---|
|
Identification |
|---|
| Name: |
octanoyl-CoA |
|---|
| Description: | An acyl-CoA(4−) that is the tetraanion of octanoyl-CoA, arising from deprotonation of phosphate and diphosphate functions. |
|---|
|
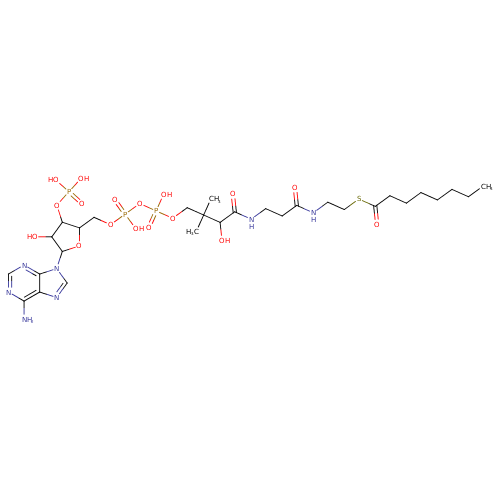
Structure |
|
|---|
| Synonyms: | -
Octanoyl-CoA (n-C8:0CoA)
-
capryloyl-CoA
|
|---|
|
Chemical Formula: |
C29H46N7O17P3S
|
|---|
| Average Molecular Weight: |
889.7 |
|---|
| Monoisotopic Molecular
Weight: |
893.2196734371 |
|---|
| InChI Key: |
KQMZYOXOBSXMII-CECATXLMSA-J |
|---|
| InChI: |
InChI=1S/C29H50N7O17P3S/c1-4-5-6-7-8-9-20(38)57-13-12-31-19(37)10-11-32-27(41)24(40)29(2,3)15-50-56(47,48)53-55(45,46)49-14-18-23(52-54(42,43)44)22(39)28(51-18)36-17-35-21-25(30)33-16-34-26(21)36/h16-18,22-24,28,39-40H,4-15H2,1-3H3,(H,31,37)(H,32,41)(H,45,46)(H,47,48)(H2,30,33,34)(H2,42,43,44)/p-4/t18-,22-,23-,24+,28-/m1/s1 |
|---|
| CAS
number: |
1264-52-4 |
|---|
| IUPAC Name: | 3'- phosphonatoadenosine 5'- phosphonatoadenosine 5'- (3- (3- {(3R)- {(3R)- 3- 3- hydroxy- hydroxy- 2,2- 2,2- dimethyl- dimethyl- 4- 4- [(3- [(3- {[2- {[2- (octanoylsulfanyl)ethyl]amino}- (octanoylsulfanyl)ethyl]amino}- 3- 3- oxopropyl)amino]- oxopropyl)amino]- 4- 4- oxobutyl} diphosphate) oxobutyl} diphosphate) |
|---|
|
Traditional IUPAC Name: |
octanoyl-coenzyme A |
|---|
| SMILES: | CCCCCCCC(=O)SCCNC(=O)CCNC(=O)C(O)C(C)(C)COP(=O)(OP(=O)(OCC1(C(OP([O-])(=O)[O-])C(O)C(O1)N3(C2(=C(C(N)=NC=N2)N=C3))))[O-])[O-] |
|---|
|
Chemical Taxonomy |
|---|
|
Taxonomy Description | This compound belongs to the class of chemical entities known as 2,3,4-saturated fatty acyl coas. These are acyl-CoAs carrying a 2,3,4-saturated fatty acyl chain. |
|---|
|
Kingdom |
Chemical entities |
|---|
| Super Class | Organic compounds |
|---|
|
Class |
Lipids and lipid-like molecules |
|---|
| Sub Class | Fatty Acyls |
|---|
|
Direct Parent |
2,3,4-saturated fatty acyl CoAs |
|---|
| Alternative Parents |
|
|---|
| Substituents |
- Coenzyme a or derivatives
- Purine ribonucleoside 3',5'-bisphosphate
- Purine ribonucleoside bisphosphate
- Purine ribonucleoside diphosphate
- Pentose phosphate
- Pentose-5-phosphate
- Ribonucleoside 3'-phosphate
- Beta amino acid or derivatives
- Glycosyl compound
- N-glycosyl compound
- Monosaccharide phosphate
- Organic pyrophosphate
- 6-aminopurine
- Pentose monosaccharide
- Imidazopyrimidine
- Purine
- Monoalkyl phosphate
- Aminopyrimidine
- Fatty amide
- Imidolactam
- Monosaccharide
- N-acyl-amine
- N-substituted imidazole
- Organic phosphoric acid derivative
- Alkyl phosphate
- Phosphoric acid ester
- Primary aromatic amine
- Pyrimidine
- Oxolane
- Azole
- Imidazole
- Heteroaromatic compound
- Amino acid or derivatives
- Thiocarboxylic acid ester
- Carboxamide group
- Carbothioic s-ester
- Secondary carboxylic acid amide
- Secondary alcohol
- Sulfenyl compound
- Thiocarboxylic acid or derivatives
- Organoheterocyclic compound
- Azacycle
- Oxacycle
- Carboxylic acid derivative
- Hydrocarbon derivative
- Alcohol
- Organic nitrogen compound
- Amine
- Organonitrogen compound
- Carbonyl group
- Organooxygen compound
- Organosulfur compound
- Organic oxygen compound
- Organopnictogen compound
- Primary amine
- Organic oxide
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework |
Aromatic heteropolycyclic compounds |
|---|
| External Descriptors |
Not Available |
|---|
|
Physical Properties |
|---|
| State: |
Solid |
|---|
| Charge: | -4 |
|---|
|
Melting point: |
Not Available |
|---|
| Experimental Properties: |
| Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties |
|
|---|
|
Biological Properties |
|---|
| Cellular Locations: |
Not Available |
|---|
| Reactions: | |
|---|
|
Pathways: |
|
|---|
|
Spectra |
|---|
| Spectra: |
|
|---|
|
References |
|---|
| References: |
- Partanen ST, Novikov DK, Popov AN, Mursula AM, Hiltunen JK, Wierenga RK: The 1.3 A crystal structure of human mitochondrial Delta3-Delta2-enoyl-CoA isomerase shows a novel mode of binding for the fatty acyl group. J Mol Biol. 2004 Sep 24;342(4):1197-208. [15351645 ]
- Schowalter DB, Matern D, Vockley J: In vitro correction of medium chain acyl CoA dehydrogenase deficiency with a recombinant adenoviral vector. Mol Genet Metab. 2005 Jun;85(2):88-95. Epub 2005 Mar 19. [15896652 ]
- Gargus JJ, Boyle K, Bocian M, Roe DS, Vianey-Saban C, Roe CR: Respiratory complex II defect in siblings associated with a symptomatic secondary block in fatty acid oxidation. J Inherit Metab Dis. 2003;26(7):659-70. [14707514 ]
- Parker AR: Binding of the human "electron transferring flavoprotein" (ETF) to the medium chain acyl-CoA dehydrogenase (MCAD) involves an arginine and histidine residue. J Enzyme Inhib Med Chem. 2003 Oct;18(5):453-62. [14692513 ]
|
|---|
| Synthesis Reference: |
Milne, K. G.; Mehlert, A.; Ferguson, M. A. J. The use of Pseudomonas acyl-CoA synthetase to form acyl-CoAs from dicarboxylic fatty acids. Biochimica et Biophysica Acta, Molecular and Cell Biology of Lipids (2001), 1531(1-2), 1-3. |
|---|
| Material Safety Data Sheet (MSDS) |
Not Available |
|---|
|
Links |
|---|
| External Links: |
|
|---|