|
Record Information |
|---|
| Version |
1.0 |
|---|
| Update Date |
1/22/2018 11:54:54 AM |
|---|
|
Metabolite ID | PAMDB004259 |
|---|
|
Identification |
|---|
| Name: |
L-Lysyl-tRNA |
|---|
| Description: | L-Lysyl-tRNA is an intermediate in tRNA charging pathway in E.coli. It is a product for the enzyme lysyl-tRNA synthetase which catalyzes the reaction ATP + L-lysine + tRNALys -> AMP + diphosphate + L-lysyl-tRNALys (BioCyc class: Charged-LYS-tRNAs). |
|---|
|
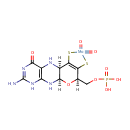
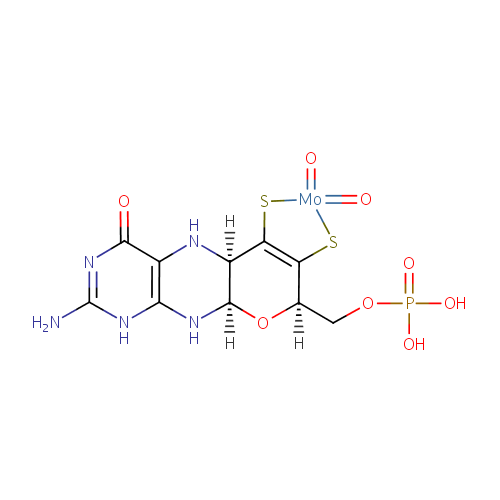
Structure |
|
|---|
| Synonyms: | - moco (dioxyo)
- Molybdenum cofactor
- Molybdenum cofactor (dioxyo)
- Molybdopterin
- Molybdopterin cofactor
- MoO2(OH)DTPP-MP
- Pterin molybdenum cofactor
|
|---|
|
Chemical Formula: |
C10H16MoN5O8PS2 |
|---|
| Average Molecular Weight: |
525.31 |
|---|
| Monoisotopic Molecular
Weight: |
526.923197 |
|---|
| InChI Key: |
VUKICSJFFDCESC-UHFFFAOYSA-L |
|---|
| InChI: | InChI=1S/C10H14N5O6PS2.Mo.2H2O/c11-10-14-7-4(8(16)15-10)12-3-6(24)5(23)2(21-9(3)13-7)1-20-22(17,18)19;;;/h2-3,9,12,23-24H,1H2,(H2,17,18,19)(H4,11,13,14,15,16);;2*1H2/q;+2;;/p-2 |
|---|
| CAS
number: |
Not Available |
|---|
| IUPAC Name: | {[(1R,10R,16R)-5-amino-7,13,13-trioxo-17-oxa-12,14-dithia-2,4,6,9-tetraaza-13-molybdatetracyclo[8.7.0.0?,??0??,???heptadeca-3(8),5,11(15)-trien-16-yl]methoxy}phosphonic acid |
|---|
|
Traditional IUPAC Name: |
[(1R,10R,16R)-5-amino-7,13,13-trioxo-17-oxa-12,14-dithia-2,4,6,9-tetraaza-13-molybdatetracyclo[8.7.0.0?,??0??,???heptadeca-3(8),5,11(15)-trien-16-yl]methoxyphosphonic acid |
|---|
| SMILES: | O.O.[Mo++].[H]C1(COP(O)([O-])=O)OC2([H])NC3=C(NC2([H])C(S)=C1S)C([O-])=NC(=N)N3 |
|---|
|
Chemical Taxonomy |
|---|
|
Taxonomy Description | Not Available |
|---|
|
Kingdom |
Not Available |
|---|
| Super Class | Not Available |
|---|
|
Class |
Not Available |
|---|
| Sub Class | Not Available |
|---|
|
Direct Parent |
Not Available |
|---|
| Alternative Parents |
Not Available |
|---|
| Substituents |
Not Available |
|---|
| Molecular Framework |
Not Available |
|---|
| External Descriptors |
Not Available |
|---|
|
Physical Properties |
|---|
| State: |
Not Available |
|---|
| Charge: | -2 |
|---|
|
Melting point: |
Not Available |
|---|
| Experimental Properties: |
|
|---|
| Predicted Properties |
|
|---|
|
Biological Properties |
|---|
| Cellular Locations: |
Cytoplasm |
|---|
| Reactions: | |
|---|
|
Pathways: |
|
|---|
|
Spectra |
|---|
| Spectra: |
|
|---|
|
References |
|---|
| References: |
Not Available |
|---|
| Synthesis Reference: |
Not Available |
|---|
| Material Safety Data Sheet (MSDS) |
Not Available |
|---|
|
Links |
|---|
| External Links: |
| Resource | Link |
|---|
| CHEBI ID | 71306 | | HMDB ID | Not Available | | Pubchem Compound ID | 56928099 | | Kegg ID | C01931 | | ChemSpider ID | 28533522 | | Wikipedia ID | Not Available | | BioCyc ID | Not Available |
|
|---|