|
Record Information |
|---|
| Version |
1.0 |
|---|
| Update Date |
1/22/2018 11:54:54 AM |
|---|
|
Metabolite ID | PAMDB002045 |
|---|
|
Identification |
|---|
| Name: |
Dextrin |
|---|
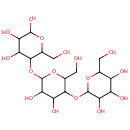
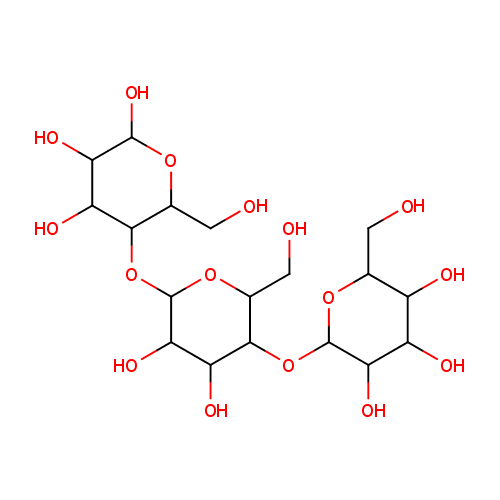
| Description: | Dextrins are a group of low-molecular-weight carbohydrates produced by the hydrolysis of starch or glycogen. Dextrins are mixtures of polymers of D-glucose units linked by -(1_4) or -(1_6) glycosidic bonds. The cyclical dextrins are known as cyclodextrins. They are formed by enzymatic degradation of starch by certain bacteria, for example, Bacillus macerans. Cyclodextrins have toroidal structures formed by 6-8 glucose residues. (Wikipedia) In Pseudomonas aeruginosa metabolism, the enzyme alpha-amylase (EC 3.2.1.1) catalyzes the breakdown of glycogen into dextrin. (KEGG) |
|---|
|
Structure |
|
|---|
| Synonyms: | - British gum
- Caloreen
- Corn dextrin
- Crystal gum
- Dextrid
- Dextrina Bianca
- Dextrine
- Dextrins
- Fortodex
|
|---|
|
Chemical Formula: |
C18H32O16 |
|---|
| Average Molecular Weight: |
504.4371 |
|---|
| Monoisotopic Molecular
Weight: |
504.169034976 |
|---|
| InChI Key: |
FYGDTMLNYKFZSV-MRCIVHHJSA-N |
|---|
| InChI: | InChI=1S/C18H32O16/c19-1-4-7(22)8(23)12(27)17(31-4)34-15-6(3-21)32-18(13(28)10(15)25)33-14-5(2-20)30-16(29)11(26)9(14)24/h4-29H,1-3H2/t4-,5-,6-,7-,8+,9-,10-,11-,12-,13-,14-,15-,16+,17?,18?/m1/s1 |
|---|
| CAS
number: |
9004-53-9 |
|---|
| IUPAC Name: | 2-{[4,5-dihydroxy-2-(hydroxymethyl)-6-{[4,5,6-trihydroxy-2-(hydroxymethyl)oxan-3-yl]oxy}oxan-3-yl]oxy}-6-(hydroxymethyl)oxane-3,4,5-triol |
|---|
|
Traditional IUPAC Name: |
amylose |
|---|
| SMILES: | OC[C@H]1OC(O[C@H]2[C@H](O)[C@@H](O)C(O[C@H]3[C@H](O)[C@@H](O)[C@@H](O)O[C@@H]3CO)O[C@@H]2CO)[C@H](O)[C@@H](O)[C@@H]1O |
|---|
|
Chemical Taxonomy |
|---|
|
Taxonomy Description | This compound belongs to the class of organic compounds known as oligosaccharides. These are carbohydrates made up of 3 to 10 monosaccharide units linked to each other through glycosidic bonds. |
|---|
|
Kingdom |
Organic compounds |
|---|
| Super Class | Organooxygen compounds |
|---|
|
Class |
Carbohydrates and carbohydrate conjugates |
|---|
| Sub Class | Oligosaccharides |
|---|
|
Direct Parent |
Oligosaccharides |
|---|
| Alternative Parents |
|
|---|
| Substituents |
- Oligosaccharide
- O-glycosyl compound
- Glycosyl compound
- Secondary alcohol
- Hemiacetal
- 1,2-diol
- Oxacycle
- Organoheterocyclic compound
- Polyol
- Acetal
- Hydrocarbon derivative
- Primary alcohol
- Alcohol
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework |
Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors |
|
|---|
|
Physical Properties |
|---|
| State: |
Solid |
|---|
| Charge: | 0 |
|---|
|
Melting point: |
Not Available |
|---|
| Experimental Properties: |
|
|---|
| Predicted Properties |
|
|---|
|
Biological Properties |
|---|
| Cellular Locations: |
Cytoplasm |
|---|
| Reactions: | |
|---|
|
Pathways: |
|
|---|
|
Spectra |
|---|
| Spectra: |
| Spectrum Type | Description | Splash Key | |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-06vu-0509620000-d5199f4e8a350b10f5a9 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0059-0809100000-9bba0a873c235685e755 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-01q9-1912000000-47259f8b59f9be52f363 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-06vu-0509620000-d5199f4e8a350b10f5a9 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0059-0809100000-9bba0a873c235685e755 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-01q9-1912000000-47259f8b59f9be52f363 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-06vu-0509620000-d5199f4e8a350b10f5a9 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0059-0809100000-9bba0a873c235685e755 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-01q9-1912000000-47259f8b59f9be52f363 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0udr-0536960000-0aea29e0372e2dfb2ff8 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00bi-2925300000-fdb113a75d83b27864c6 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-3921000000-8faa9fe46329a3130ea9 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0udr-0536960000-0aea29e0372e2dfb2ff8 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00bi-2925300000-fdb113a75d83b27864c6 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-3921000000-8faa9fe46329a3130ea9 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0udr-0536960000-0aea29e0372e2dfb2ff8 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00bi-2925300000-fdb113a75d83b27864c6 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-3921000000-8faa9fe46329a3130ea9 | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available |
|---|
|
|---|
|
References |
|---|
| References: |
- Yannai, Shmuel. (2004) Dictionary of food compounds with CD-ROM: Additives, flavors, and ingredients. Boca Raton: Chapman & Hall/CRC.
|
|---|
| Synthesis Reference: |
Not Available |
|---|
| Material Safety Data Sheet (MSDS) |
Download (PDF) |
|---|
|
Links |
|---|
| External Links: |
|
|---|