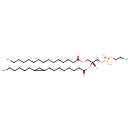
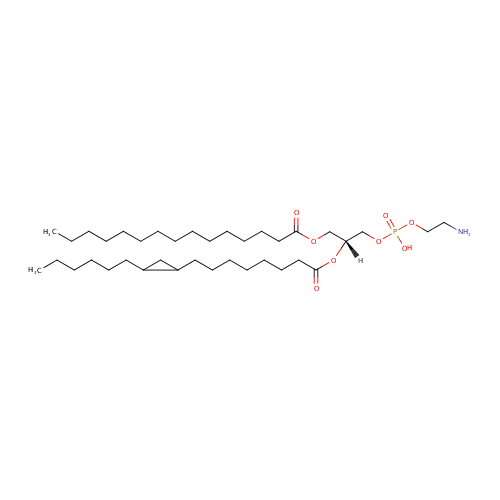
| InChI: | InChI=1S/C37H72NO8P/c1-3-5-7-9-10-11-12-13-14-15-18-22-26-36(39)43-31-35(32-45-47(41,42)44-29-28-38)46-37(40)27-23-19-16-17-21-25-34-30-33(34)24-20-8-6-4-2/h33-35H,3-32,38H2,1-2H3,(H,41,42)/t33?,34?,35-/m1/s1 |
|---|
| References: |
- Kanehisa, M., Goto, S., Sato, Y., Furumichi, M., Tanabe, M. (2012). "KEGG for integration and interpretation of large-scale molecular data sets." Nucleic Acids Res 40:D109-D114. Pubmed: 22080510
- Keseler, I. M., Collado-Vides, J., Santos-Zavaleta, A., Peralta-Gil, M., Gama-Castro, S., Muniz-Rascado, L., Bonavides-Martinez, C., Paley, S., Krummenacker, M., Altman, T., Kaipa, P., Spaulding, A., Pacheco, J., Latendresse, M., Fulcher, C., Sarker, M., Shearer, A. G., Mackie, A., Paulsen, I., Gunsalus, R. P., Karp, P. D. (2011). "EcoCyc: a comprehensive database of Escherichia coli biology." Nucleic Acids Res 39:D583-D590. Pubmed: 21097882
- Oursel, D., Loutelier-Bourhis, C., Orange, N., Chevalier, S., Norris, V., Lange, C. M. (2007). "Lipid composition of membranes of Escherichia coli by liquid chromatography/tandem mass spectrometry using negative electrospray ionization." Rapid Commun Mass Spectrom 21:1721-1728. Pubmed: 17477452
- Yurtsever D. (2007). Fatty acid methyl ester profiling of Enterococcus and Esherichia coli for microbial source tracking. M.sc. Thesis. Villanova University: U.S.A
|
|---|