|
Record Information |
|---|
| Version |
1.0 |
|---|
| Update Date |
1/22/2018 11:54:54 AM |
|---|
|
Metabolite ID | PAMDB001875 |
|---|
|
Identification |
|---|
| Name: |
2,3-Dihydroxyisovaleric acid |
|---|
| Description: | 2,3-dihydroxyisovaleric acid (or 2,3-Dihydroxy-3-methylbutanoate) is invovled in branched chain amino acid biosynthesis. It is a substrate for Ketol-acid reductoisomerase (ilvC). This enzyme catalyzes the reaction: (R)-2,3-dihydroxy-3-methylbutanoate + NADP+ = (S)-2-hydroxy-2-methyl-3-oxobutanoate + NADPH. |
|---|
|
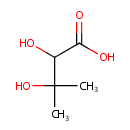
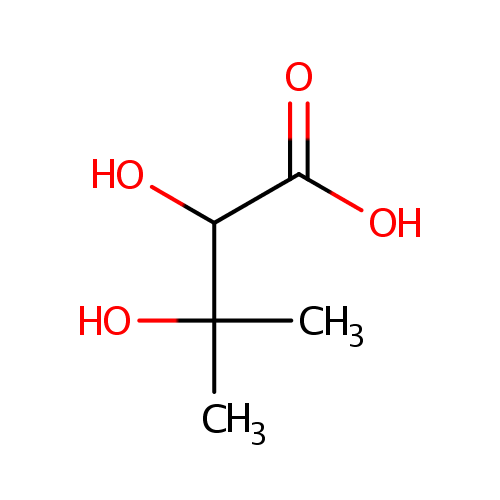
Structure |
|
|---|
| Synonyms: | - 2,3-Dihydroxy-3-methylbutanoate
- 2,3-Dihydroxy-3-methylbutanoic acid
- 2,3-Dihydroxy-isovalerate
- 2,3-Dihydroxy-isovaleric acid
- 2,3-Dihydroxyisovalerate
- 2,3-DIV
- a,b-Dihydroxyisovalerate
- a,b-Dihydroxyisovaleric acid
- Alpha,beta-Dihydroxyisovalerate
- Alpha,beta-Dihydroxyisovaleric acid
- α,β-Dihydroxyisovalerate
- α,β-Dihydroxyisovaleric acid
|
|---|
|
Chemical Formula: |
C5H10O4 |
|---|
| Average Molecular Weight: |
134.1305 |
|---|
| Monoisotopic Molecular
Weight: |
134.057908808 |
|---|
| InChI Key: |
JTEYKUFKXGDTEU-UHFFFAOYSA-N |
|---|
| InChI: | InChI=1S/C5H10O4/c1-5(2,9)3(6)4(7)8/h3,6,9H,1-2H3,(H,7,8) |
|---|
| CAS
number: |
1756-18-9 |
|---|
| IUPAC Name: | 2,3-dihydroxy-3-methylbutanoic acid |
|---|
|
Traditional IUPAC Name: |
2,3-dihydroxyisovaleric acid |
|---|
| SMILES: | CC(C)(O)C(O)C(O)=O |
|---|
|
Chemical Taxonomy |
|---|
|
Taxonomy Description | This compound belongs to the class of organic compounds known as hydroxy fatty acids. These are fatty acids in which the chain bears a hydroxyl group. |
|---|
|
Kingdom |
Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
|
Class |
Fatty Acyls |
|---|
| Sub Class | Fatty acids and conjugates |
|---|
|
Direct Parent |
Hydroxy fatty acids |
|---|
| Alternative Parents |
|
|---|
| Substituents |
- Hydroxy fatty acid
- Methyl-branched fatty acid
- Short-chain hydroxy acid
- Branched fatty acid
- Beta-hydroxy acid
- Monosaccharide
- Hydroxy acid
- Alpha-hydroxy acid
- Tertiary alcohol
- Secondary alcohol
- 1,2-diol
- Monocarboxylic acid or derivatives
- Carboxylic acid
- Carboxylic acid derivative
- Hydrocarbon derivative
- Organooxygen compound
- Carbonyl group
- Alcohol
- Aliphatic acyclic compound
|
|---|
| Molecular Framework |
Aliphatic acyclic compounds |
|---|
| External Descriptors |
Not Available |
|---|
|
Physical Properties |
|---|
| State: |
Not Available |
|---|
| Charge: | -1 |
|---|
|
Melting point: |
Not Available |
|---|
| Experimental Properties: |
|
|---|
| Predicted Properties |
|
|---|
|
Biological Properties |
|---|
| Cellular Locations: |
Cytoplasm |
|---|
| Reactions: | |
|---|
|
Pathways: |
- Metabolic pathways pae01100
- Pantothenate and CoA biosynthesis pae00770
- Valine, leucine and isoleucine biosynthesis pae00290
|
|---|
|
Spectra |
|---|
| Spectra: |
|
|---|
|
References |
|---|
| References: |
- Winder, C. L., Dunn, W. B., Schuler, S., Broadhurst, D., Jarvis, R., Stephens, G. M., Goodacre, R. (2008). "Global metabolic profiling of Escherichia coli cultures: an evaluation of methods for quenching and extraction of intracellular metabolites." Anal Chem 80:2939-2948. Pubmed: 18331064
- Yurtsever D. (2007). Fatty acid methyl ester profiling of Enterococcus and Esherichia coli for microbial source tracking. M.sc. Thesis. Villanova University: U.S.A
|
|---|
| Synthesis Reference: |
Not Available |
|---|
| Material Safety Data Sheet (MSDS) |
Not Available |
|---|
|
Links |
|---|
| External Links: |
| Resource | Link |
|---|
| CHEBI ID | 15689 | | HMDB ID | Not Available | | Pubchem Compound ID | 677 | | Kegg ID | C04039 | | ChemSpider ID | 657 | | Wikipedia ID | Not Available | | BioCyc ID | Not Available |
|
|---|