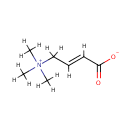
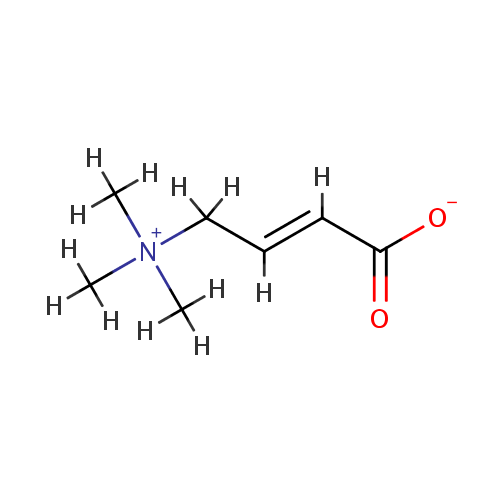
Crotonobetaine (PAMDB001723)
| Record Information | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 1.0 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 1/22/2018 11:54:54 AM | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolite ID | PAMDB001723 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | Crotonobetaine | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description: | Crotonobetaine is a member of the chemical class known as Quaternary Ammonium Salts. These are compounds containing positively charged polyatomic ion of the structure NR4+, R being an alkyl group or an aryl groupCrotonobetaine is invovled in Carnitine metabolism, and ABC transporters. Crotonobetaine is involved in carnitine metabolism. Carnitine dehydratase from Pseudomonas aeruginosa O44 K74 is an inducible enzyme detectable in cells grown anaerobically in the presence of L-(-)-carnitine or crotonobetaine. (PMID 8188598). | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C7H13NO2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Weight: | 143.1836 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Molecular Weight: | 143.094628665 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | GUYHPGUANSLONG-SNAWJCMRSA-N | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C7H13NO2/c1-8(2,3)6-4-5-7(9)10/h4-5H,6H2,1-3H3/b5-4+ | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | 927-89-9 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | (2E)-4-(trimethylazaniumyl)but-2-enoate | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | crotonobetaine | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | [H]\C(=C(\[H])C([H])([H])[N+](C([H])([H])[H])(C([H])([H])[H])C([H])([H])[H])C([O-])=O | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Taxonomy Description | This compound belongs to the class of organic compounds known as straight chain fatty acids. These are fatty acids with a straight aliphatic chain. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Lipids and lipid-like molecules | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Fatty Acyls | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Fatty acids and conjugates | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Straight chain fatty acids | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aliphatic acyclic compounds | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | 0 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | Adenosine triphosphate + Water + Crotonobetaine > ADP + Crotonobetaine + Hydrogen ion + Phosphate Adenosine triphosphate + Water + Crotonobetaine > ADP + Crotonobetaine + Hydrogen ion + Phosphate Adenosine triphosphate + Coenzyme A + Crotonobetaine > ADP + Crotonobetainyl-CoA + Phosphate Carnitine + Crotonobetainyl-CoA <> L-Carnitinyl-CoA + Crotonobetaine (E)-4-(Trimethylammonio)but-2-enoyl-CoA + L-Carnitine + 4-Trimethylammoniobutanoyl-CoA <> Crotonobetaine + L-Carnitinyl-CoA + gamma-Butyrobetaine | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways: | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Enzymes
- General function:
- Involved in nucleotide binding
- Specific function:
- Involved in a multicomponent binding-protein-dependent transport system for glycine betaine/L-proline
- Gene Name:
- proV
- Locus Tag:
- PA5094
- Molecular weight:
- 30.7 kDa