3-Sulfopyruvic acid (PAMDB001422)
| Record Information | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 1.0 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 1/22/2018 11:54:54 AM | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolite ID | PAMDB001422 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
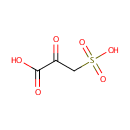
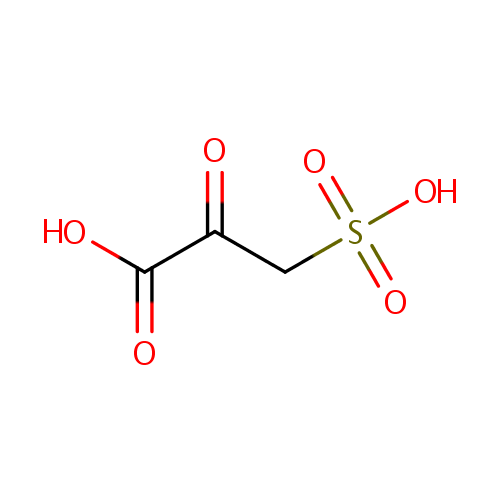
| Name: | 3-Sulfopyruvic acid | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description: | 3-Sulfopyruvic acid is the product of the transamination of cysteinesulfonate in a reaction catalyzed by aspartate aminotransferase. 3-sulfopyruvic acid is stable and is reduced by malate dehydrogenase to beta-sulfolactate. Cysteinesulfonate, 3-sulfopyruvic acid, and beta-sulfolactate are reversibly interconverted in vivo. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C3H4O6S | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Weight: | 168.125 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Molecular Weight: | 167.97285855 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | BUTHMSUEBYPMKJ-UHFFFAOYSA-N | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C3H4O6S/c4-2(3(5)6)1-10(7,8)9/h1H2,(H,5,6)(H,7,8,9) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | 98022-26-5 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | 2-oxo-3-sulfopropanoic acid | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | sulfopyruvate | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | OC(=O)C(=O)CS(O)(=O)=O | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Taxonomy Description | This compound belongs to the class of organic compounds known as alpha-keto acids and derivatives. These are organic compounds containing an aldehyde substituted with a keto group on the adjacent carbon. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organic acids and derivatives | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Keto acids and derivatives | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Alpha-keto acids and derivatives | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Alpha-keto acids and derivatives | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aliphatic acyclic compounds | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Solid | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | -2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways: | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Enzymes
- General function:
- Involved in transporter activity
- Specific function:
- Part of a binding-protein-dependent transport system for aliphatic sulfonates. Probably responsible for the translocation of the substrate across the membrane
- Gene Name:
- ssuC
- Locus Tag:
- PA3443
- Molecular weight:
- 28.5 kDa
Transporters
- General function:
- Involved in transporter activity
- Specific function:
- Part of a binding-protein-dependent transport system for aliphatic sulfonates. Probably responsible for the translocation of the substrate across the membrane
- Gene Name:
- ssuC
- Locus Tag:
- PA3443
- Molecular weight:
- 28.5 kDa