|
Record Information |
|---|
| Version |
1.0 |
|---|
| Update Date |
1/22/2018 11:54:54 AM |
|---|
|
Metabolite ID | PAMDB001368 |
|---|
|
Identification |
|---|
| Name: |
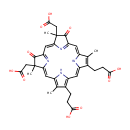
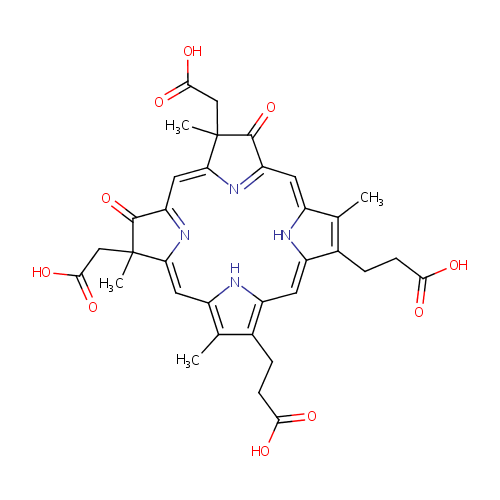
Heme D |
|---|
| Description: | Heme D is a member of the chemical class known as tetrapyrroles and Derivatives. These are polycyclic aromatic compounds containing four pyrrole rings joined by one-carbon units linking position 2 of one pyrrole ring to position 5 of the next. (inferred from compound structure). A heme (American English) or haem (British English) is a prosthetic group that consists of an iron atom contained in the center of a large heterocyclic organic ring called a porphyrin. Heme D is a derivative of heme B, but in which the propionic acid side chain at the carbon of position 6, which is also hydroxylated, forms a ??-spirolactone. Ring III is also hydroxylated at position 5, in a conformation trans to the new lactone group. Heme D is the site for oxygen reduction to water of many types of bacteria at low oxygen tension. In Pseudomonas aeruginosa, heme D is formed from protoheme in the interior of the catalase hydroperoxidase II (HPII) molecule through a self-catalyzed reaction. [PMID: 8621527] |
|---|
|
Structure |
|
|---|
| Synonyms: | Not Available |
|---|
|
Chemical Formula: |
C34H34N4O10 |
|---|
| Average Molecular Weight: |
658.6546 |
|---|
| Monoisotopic Molecular
Weight: |
658.227493328 |
|---|
| InChI Key: |
ZAKGXCNMJBQQNZ-CXJCYTKDSA-N |
|---|
| InChI: | InChI=1S/C34H34N4O10/c1-15-17(5-7-27(39)40)21-10-22-18(6-8-28(41)42)16(2)20(36-22)11-25-33(3,13-29(43)44)32(48)24(38-25)12-26-34(4,14-30(45)46)31(47)23(37-26)9-19(15)35-21/h9-12,35-36H,5-8,13-14H2,1-4H3,(H,39,40)(H,41,42)(H,43,44)(H,45,46)/b19-9-,20-11-,21-10-,22-10-,23-9-,24-12-,25-11-,26-12- |
|---|
| CAS
number: |
60318-31-2 |
|---|
| IUPAC Name: | 3-[20-(2-carboxyethyl)-10,15-bis(carboxymethyl)-5,10,15,19-tetramethyl-9,14-dioxo-21,22,23,24-tetraazapentacyclo[16.2.1.1?,??1????.1??,???tetracosa-1(20),2,4,6,8(23),11,13(22),16,18-nonaen-4-yl]propanoic acid |
|---|
|
Traditional IUPAC Name: |
3-[20-(2-carboxyethyl)-10,15-bis(carboxymethyl)-5,10,15,19-tetramethyl-9,14-dioxo-21,22,23,24-tetraazapentacyclo[16.2.1.1?,??1????.1??,???tetracosa-1(20),2,4,6,8(23),11,13(22),16,18-nonaen-4-yl]propanoic acid |
|---|
| SMILES: | CC1=C(CCC(O)=O)/C2=C/C3=C(CCC(O)=O)C(C)=C(N3)\C=C3/N=C(/C=C4\N=C(\C=C\1/N\2)C(=O)C4(C)CC(O)=O)C(=O)C3(C)CC(O)=O |
|---|
|
Chemical Taxonomy |
|---|
|
Taxonomy Description | This compound belongs to the class of organic compounds known as tetrapyrroles and derivatives. These are polycyclic aromatic compounds containing four pyrrole rings joined by one-carbon units linking position 2 of one pyrrole ring to position 5 of the next. |
|---|
|
Kingdom |
Organic compounds |
|---|
| Super Class | Organoheterocyclic compounds |
|---|
|
Class |
Tetrapyrroles and derivatives |
|---|
| Sub Class | Not Available |
|---|
|
Direct Parent |
Tetrapyrroles and derivatives |
|---|
| Alternative Parents |
|
|---|
| Substituents |
- Tetrapyrrole skeleton
- Tetracarboxylic acid or derivatives
- Aryl alkyl ketone
- Aryl ketone
- Substituted pyrrole
- Heteroaromatic compound
- Pyrrole
- Ketone
- Azacycle
- Carboxylic acid
- Carboxylic acid derivative
- Hydrocarbon derivative
- Organooxygen compound
- Organonitrogen compound
- Carbonyl group
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework |
Aromatic heteropolycyclic compounds |
|---|
| External Descriptors |
Not Available |
|---|
|
Physical Properties |
|---|
| State: |
Not Available |
|---|
| Charge: | -4 |
|---|
|
Melting point: |
Not Available |
|---|
| Experimental Properties: |
|
|---|
| Predicted Properties |
|
|---|
|
Biological Properties |
|---|
| Cellular Locations: |
Membrane |
|---|
| Reactions: | |
|---|
|
Pathways: |
|
|---|
|
Spectra |
|---|
| Spectra: |
|
|---|
|
References |
|---|
| References: |
- Keseler, I. M., Collado-Vides, J., Santos-Zavaleta, A., Peralta-Gil, M., Gama-Castro, S., Muniz-Rascado, L., Bonavides-Martinez, C., Paley, S., Krummenacker, M., Altman, T., Kaipa, P., Spaulding, A., Pacheco, J., Latendresse, M., Fulcher, C., Sarker, M., Shearer, A. G., Mackie, A., Paulsen, I., Gunsalus, R. P., Karp, P. D. (2011). "EcoCyc: a comprehensive database of Escherichia coli biology." Nucleic Acids Res 39:D583-D590. Pubmed: 21097882
- Murshudov, G. N., Grebenko, A. I., Barynin, V., Dauter, Z., Wilson, K. S., Vainshtein, B. K., Melik-Adamyan, W., Bravo, J., Ferran, J. M., Ferrer, J. C., Switala, J., Loewen, P. C., Fita, I. (1996). "Structure of the heme d of Penicillium vitale and Escherichia coli catalases." J Biol Chem 271:8863-8868. Pubmed: 8621527
- van der Werf, M. J., Overkamp, K. M., Muilwijk, B., Coulier, L., Hankemeier, T. (2007). "Microbial metabolomics: toward a platform with full metabolome coverage." Anal Biochem 370:17-25. Pubmed: 17765195
|
|---|
| Synthesis Reference: |
Not Available |
|---|
| Material Safety Data Sheet (MSDS) |
Not Available |
|---|
|
Links |
|---|
| External Links: |
| Resource | Link |
|---|
| CHEBI ID | Not Available | | HMDB ID | Not Available | | Pubchem Compound ID | 3582 | | Kegg ID | Not Available | | ChemSpider ID | 3456 | | Wikipedia ID | Not Available | | BioCyc ID | HEME_D | | EcoCyc ID | HEME_D |
|
|---|