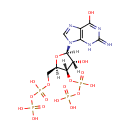
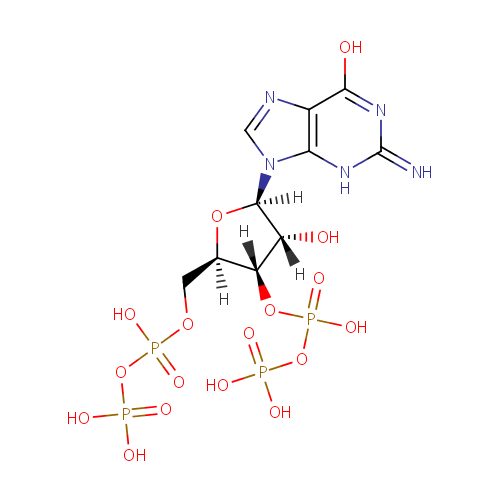
Guanosine 3',5'-bis(diphosphate) (PAMDB001005)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 1.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 1/22/2018 11:54:54 AM | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolite ID | PAMDB001005 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | Guanosine 3',5'-bis(diphosphate) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description: | (p)ppGpp, guanosine pentaphosphate or tetraphosphate is an alarmone which is involved in the stringent response in bacteria, causing the inhibition of RNA synthesis when there is a shortage of amino acids present. This causes translation to decrease and the amino acids present are therefore conserved. (p)ppGpp is an effector molecule which is produced as a result of amino acid starvation within a bacterial cell. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C10H17N5O17P4 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Weight: | 603.1603 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Molecular Weight: | 602.956990191 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | BUFLLCUFNHESEH-UUOKFMHZSA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C10H17N5O17P4/c11-10-13-7-4(8(17)14-10)12-2-15(7)9-5(16)6(30-36(26,27)32-34(21,22)23)3(29-9)1-28-35(24,25)31-33(18,19)20/h2-3,5-6,9,16H,1H2,(H,24,25)(H,26,27)(H2,18,19,20)(H2,21,22,23)(H3,11,13,14,17)/t3-,5-,6-,9-/m1/s1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | 33503-72-9 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | {[hydroxy({[(2R,3S,4R,5R)-4-hydroxy-2-({[hydroxy(phosphonooxy)phosphoryl]oxy}methyl)-5-(6-hydroxy-2-imino-3,9-dihydro-2H-purin-9-yl)oxolan-3-yl]oxy})phosphoryl]oxy}phosphonic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | {hydroxy[(2R,3S,4R,5R)-4-hydroxy-2-({[hydroxy(phosphonooxy)phosphoryl]oxy}methyl)-5-(6-hydroxy-2-imino-3H-purin-9-yl)oxolan-3-yl]oxyphosphoryl}oxyphosphonic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | [H][C@]1(COP(O)(=O)OP(O)(O)=O)O[C@@]([H])(N2C=NC3=C2NC(=N)N=C3O)[C@]([H])(O)[C@]1([H])OP(O)(=O)OP(O)(O)=O | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Taxonomy Description | This compound belongs to the class of organic compounds known as purine ribonucleoside diphosphates. These are purine ribobucleotides with diphosphate group linked to the ribose moiety. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Nucleosides, nucleotides, and analogues | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Purine nucleotides | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Purine ribonucleotides | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Purine ribonucleoside diphosphates | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aromatic heteropolycyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | -5 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | Adenosine triphosphate + Guanosine diphosphate > Adenosine monophosphate + Hydrogen ion + Guanosine 3',5'-bis(diphosphate) Water + Guanosine 3',5'-bis(diphosphate) <> Guanosine diphosphate + Pyrophosphate Guanosine 3'-diphosphate 5'-triphosphate + Water <> Guanosine 3',5'-bis(diphosphate) + Phosphate Adenosine triphosphate + Guanosine diphosphate > Adenosine monophosphate + Guanosine 3',5'-bis(diphosphate) Guanosine 3',5'-bis(diphosphate) + Water > Hydrogen ion + Pyrophosphate + Guanosine diphosphate Water + Guanosine 3'-diphosphate 5'-triphosphate > Hydrogen ion + Phosphate + Guanosine 3',5'-bis(diphosphate) Guanosine 3'-diphosphate 5'-triphosphate + Water > Guanosine 3',5'-bis(diphosphate) + Inorganic phosphate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||