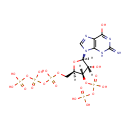
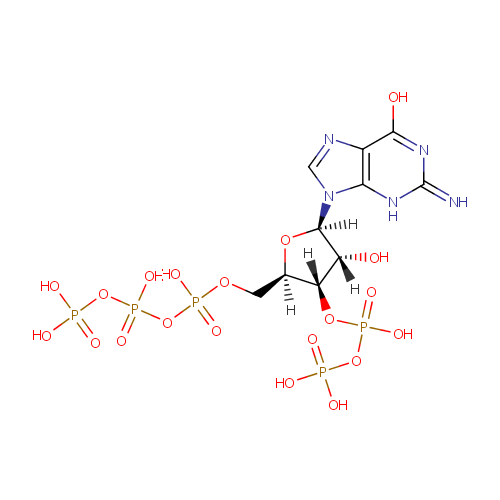
Guanosine 3'-diphosphate 5'-triphosphate (PAMDB001003)
| Record Information | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 1.0 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 1/22/2018 11:54:54 AM | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolite ID | PAMDB001003 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | Guanosine 3'-diphosphate 5'-triphosphate | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description: | Guanosine 3'-diphosphate 5'-triphosphate is a member of the chemical class known as Purine Ribonucleoside 3',5'-Bisphosphates. These are purine ribobucleotides with one phosphate group attached to 3' and 5' hydroxyl groups of the ribose moiety. The hallmark of the stringent response is the accumulation of guanosine tetra- (ppGpp) and pentaphosphates (pppGpp), which probably bind RNA polymerase to regulate gene expression at certain promoters. (PMID 11282471) Guanosine 5'-triphosphate 3'-diphosphate (pppGpp) and guanosine 5'-diphosphate 3'-diphosphate (ppGpp) are specifically degraded by a manganese-dependent pyrophosphorylase present in spoT+ but not in spoT- strains of Pseudomonas aeruginosa, indicating that the enzyme is the spoT gene product. (PMID 365225) The kinetics of the GTP level during starvation suggests that GTP is a precursor of pppGpp. (PMID 793688) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C10H18N5O20P5 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Weight: | 683.1402 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Molecular Weight: | 682.923320601 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | KCPMACXZAITQAX-UUOKFMHZSA-N | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C10H18N5O20P5/c11-10-13-7-4(8(17)14-10)12-2-15(7)9-5(16)6(32-39(26,27)33-36(18,19)20)3(31-9)1-30-38(24,25)35-40(28,29)34-37(21,22)23/h2-3,5-6,9,16H,1H2,(H,24,25)(H,26,27)(H,28,29)(H2,18,19,20)(H2,21,22,23)(H3,11,13,14,17)/t3-,5-,6-,9-/m1/s1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | 38918-96-6 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | {[hydroxy({[(2R,3S,4R,5R)-4-hydroxy-2-({[hydroxy({[hydroxy(phosphonooxy)phosphoryl]oxy})phosphoryl]oxy}methyl)-5-(6-hydroxy-2-imino-3,9-dihydro-2H-purin-9-yl)oxolan-3-yl]oxy})phosphoryl]oxy}phosphonic acid | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | {hydroxy[(2R,3S,4R,5R)-4-hydroxy-5-(6-hydroxy-2-imino-3H-purin-9-yl)-2-[({hydroxy[hydroxy(phosphonooxy)phosphoryl]oxyphosphoryl}oxy)methyl]oxolan-3-yl]oxyphosphoryl}oxyphosphonic acid | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | [H][C@]1(COP(O)(=O)OP(O)(=O)OP(O)(O)=O)O[C@@]([H])(N2C=NC3=C2NC(=N)N=C3O)[C@]([H])(O)[C@]1([H])OP(O)(=O)OP(O)(O)=O | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Taxonomy Description | This compound belongs to the class of organic compounds known as purine ribonucleoside triphosphates. These are purine ribobucleotides with a triphosphate group linked to the ribose moiety. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Nucleosides, nucleotides, and analogues | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Purine nucleotides | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Purine ribonucleotides | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Purine ribonucleoside triphosphates | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aromatic heteropolycyclic compounds | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | -6 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | Adenosine triphosphate + Guanosine triphosphate <> Adenosine monophosphate + Guanosine 3'-diphosphate 5'-triphosphate + Hydrogen ion Guanosine 3'-diphosphate 5'-triphosphate + Water > Guanosine triphosphate + Pyrophosphate Adenosine triphosphate + Guanosine triphosphate <> Adenosine monophosphate + Guanosine 3'-diphosphate 5'-triphosphate Guanosine 3'-diphosphate 5'-triphosphate + Water <> Guanosine 3',5'-bis(diphosphate) + Phosphate Water + Guanosine 3'-diphosphate 5'-triphosphate > Hydrogen ion + Phosphate + Guanosine 3',5'-bis(diphosphate) Guanosine 3'-diphosphate 5'-triphosphate + Water > Hydrogen ion + Guanosine triphosphate + Pyrophosphate Guanosine 3'-diphosphate 5'-triphosphate + Water > Guanosine 3',5'-bis(diphosphate) + Inorganic phosphate | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||