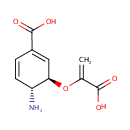
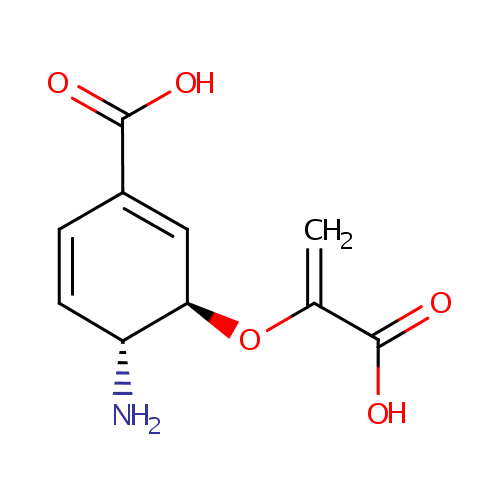
4-Amino-4-deoxychorismate (PAMDB000933)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 1.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 1/22/2018 11:54:54 AM | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolite ID | PAMDB000933 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | 4-Amino-4-deoxychorismate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description: | 4-amino-4-deoxychorismate (or ADC) can be classified as a member of the Dicarboxylic Acids and Derivatives. These are organic compounds containing exactly two carboxylic acid groups. ADC is involved in antibiotic biosynthesis. In some antibiotic producers, p-aminobenzoic acid (PABA) or its immediate precursor, 4-amino-4-deoxychorismate (ADC), is involved in primary metabolism and antibiotic biosynthesis. (PMID 19389784) In Pseudomonas aeruginosa, the production of PABA is catalyzed by the PabC protein, a beta-lyase that converts 4-amino-4-deoxychorismate (ADC)--the reaction product of the PabA and PabB enzymes--to PABA and pyruvate. (PMID 15500462) 4-Amino-4-deoxychorismate lyase (ADCL) is a member of the fold-type IV of PLP dependent enzymes that converts 4-amino-4-deoxychorismate (ADC) to p-aminobenzoate and pyruvate. (PMID 10876155) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C10H10NO5 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Weight: | 224.193 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Molecular Weight: | 224.056446006 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | OIUJHGOLFKDBSU-HTQZYQBOSA-M | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C10H11NO5/c1-5(9(12)13)16-8-4-6(10(14)15)2-3-7(8)11/h2-4,7-8H,1,11H2,(H,12,13)(H,14,15)/p-1/t7-,8-/m1/s1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | 133442-18-9 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | (3R,4R)-4-amino-3-[(1-carboxyeth-1-en-1-yl)oxy]cyclohexa-1,5-diene-1-carboxylic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | 4-amino-4-deoxychorismic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | [H][C@@]1(N)C=CC(=C[C@@]1([H])OC(=C)C([O-])=O)C(O)=O | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Taxonomy Description | This compound belongs to the class of organic compounds known as dicarboxylic acids and derivatives. These are organic compounds containing exactly two carboxylic acid groups. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organic acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Carboxylic acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Dicarboxylic acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Dicarboxylic acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aliphatic homomonocyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | -1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | Chorismate + L-Glutamine <> 4-Amino-4-deoxychorismate + L-Glutamate 4-Amino-4-deoxychorismate <> p-Aminobenzoic acid + Hydrogen ion + Pyruvic acid 4-Amino-4-deoxychorismate <> p-Aminobenzoic acid + Pyruvic acid 4-amino-4-deoxychorismate + 4-Amino-4-deoxychorismate > Pyruvic acid + Hydrogen ion + p-Aminobenzoic acid Chorismate + L-Glutamine > L-Glutamic acid + 4-amino-4-deoxychorismate + L-Glutamate + 4-Amino-4-deoxychorismate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||