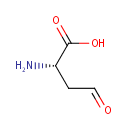
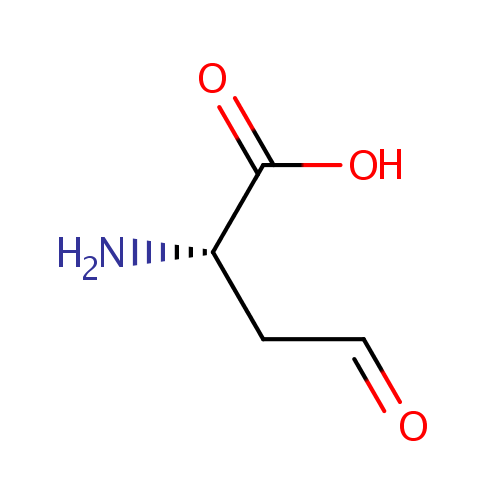
L-Aspartate-semialdehyde (PAMDB000833)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 1.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 1/22/2018 11:54:54 AM | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolite ID | PAMDB000833 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | L-Aspartate-semialdehyde | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description: | L-Aspartate-semialdehyde is involved in both the lysine biosynthesis and homoserine biosynthesis pathways. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C4H7NO3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Weight: | 117.1033 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Molecular Weight: | 117.042593095 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | HOSWPDPVFBCLSY-VKHMYHEASA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C4H7NO3/c5-3(1-2-6)4(7)8/h2-3H,1,5H2,(H,7,8)/t3-/m0/s1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | 15106-57-7 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | (2S)-2-amino-4-oxobutanoic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | L-aspartic 4-semialdehyde | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | N[C@@H](CC=O)C(O)=O | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Taxonomy Description | This compound belongs to the class of organic compounds known as d-alpha-amino acids. These are alpha amino acids which have the D-configuration of the alpha-carbon atom. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organic acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Carboxylic acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Amino acids, peptides, and analogues | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | D-alpha-amino acids | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aliphatic acyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Solid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | 0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | L-Homoserine + NADP <> L-Aspartate-semialdehyde + Hydrogen ion + NADPH L-Aspartate-semialdehyde + NADP + Phosphate <> L-Aspartyl-4-phosphate + Hydrogen ion + NADPH L-Homoserine + NAD <> L-Aspartate-semialdehyde + NADH + Hydrogen ion L-Aspartate-semialdehyde + Pyruvic acid <> 2,3-Dihydrodipicolinic acid +2 Water Pyruvic acid + L-Aspartate-semialdehyde <> Hydrogen ion + Water + 2,3-Dihydrodipicolinic acid NAD(P)<sup>+</sup> + L-Homoserine < NAD(P)H + L-Aspartate-semialdehyde + Hydrogen ion L-Homoserine + NAD(P)(+) > L-Aspartate-semialdehyde + NAD(P)H Pyruvic acid + L-Aspartate-semialdehyde <> (2S,4S)-4-Hydroxy-2,3,4,5-tetrahydrodipicolinate + Water L-Aspartyl-4-phosphate + NADPH + Hydrogen ion + NADPH > Phosphate + NADP + L-Aspartate-semialdehyde L-Aspartate-semialdehyde + Pyruvic acid > Hydrogen ion + Water + (2S,4S)-4-hydroxy-2,3,4,5-tetrahydrodipicolinate + (2S,4S)-4-Hydroxy-2,3,4,5-tetrahydrodipicolinate L-Aspartate-semialdehyde + Hydrogen ion + NADPH + NADPH > NADP + L-Homoserine + L-Homoserine | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Enzymes
- General function:
- Involved in catalytic activity
- Specific function:
- L-aspartate 4-semialdehyde + pyruvate = dihydrodipicolinate + 2 H(2)O
- Gene Name:
- dapA
- Locus Tag:
- PA1010
- Molecular weight:
- 31.4 kDa
Reactions
| L-aspartate 4-semialdehyde + pyruvate = dihydrodipicolinate + 2 H(2)O. |
- General function:
- Involved in aspartate-semialdehyde dehydrogenase activity
- Specific function:
- L-aspartate 4-semialdehyde + phosphate + NADP(+) = L-4-aspartyl phosphate + NADPH
- Gene Name:
- asd
- Locus Tag:
- PA3117
- Molecular weight:
- 40.5 kDa
Reactions
| L-aspartate 4-semialdehyde + phosphate + NADP(+) = L-4-aspartyl phosphate + NADPH. |