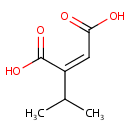
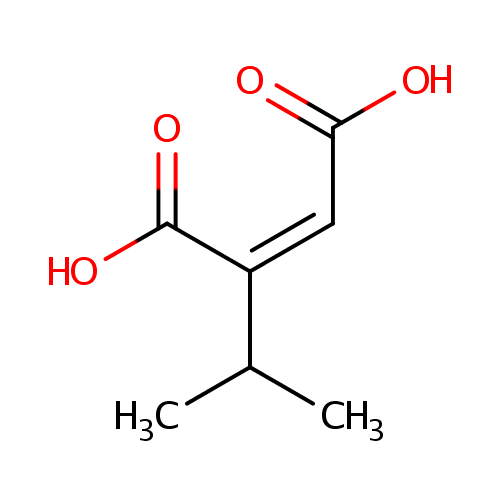
Isopropylmaleate (PAMDB000832)
| Record Information | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 1.0 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 1/22/2018 11:54:54 AM | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolite ID | PAMDB000832 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | Isopropylmaleate | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description: | Isopropylmaleate is an intermediate in branched chain amino acid metabolism (specifically L-leucine biosynthesis). It is a substrate for 3-isopropylmalate dehydratase. This enzyme catalyzes the isomerization between 2-isopropylmalate and 3-isopropylmalate, via the formation of 2-isopropylmaleate. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C7H10O4 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Weight: | 158.1519 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Molecular Weight: | 158.057908808 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | NJMGRJLQRLFQQX-HYXAFXHYSA-N | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C7H10O4/c1-4(2)5(7(10)11)3-6(8)9/h3-4H,1-2H3,(H,8,9)(H,10,11)/b5-3- | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | (2Z)-2-(propan-2-yl)but-2-enedioic acid | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | 2-isopropylmaleic acid | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | CC(C)C(=C\C(O)=O)\C(O)=O | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Taxonomy Description | This compound belongs to the class of organic compounds known as methyl-branched fatty acids. These are fatty acids with an acyl chain that has a methyl branch. Usually, they are saturated and contain only one or more methyl group. However, branches other than methyl may be present. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Lipids and lipid-like molecules | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Fatty Acyls | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Fatty acids and conjugates | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Methyl-branched fatty acids | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aliphatic acyclic compounds | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Solid | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | -2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways: | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||