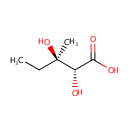
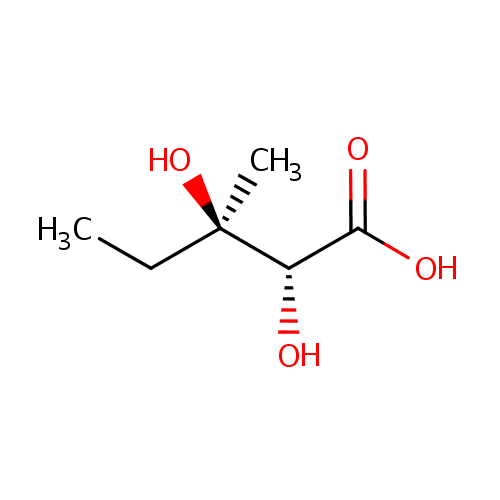
(R) 2,3-Dihydroxy-3-methylvalerate (PAMDB000822)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 1.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 1/22/2018 11:54:54 AM | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolite ID | PAMDB000822 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | (R) 2,3-Dihydroxy-3-methylvalerate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description: | (R) 2,3-Dihydroxy-methylvalerate is an intermediate in valine, leucine and isoleucine biosynthesis. The pathway of valine biosynthesis is a four-step pathway that shares all of its steps with the parallel pathway of isoleucine biosynthesis. These entwined pathways are part of the superpathway of leucine, valine, and isoleucine biosynthesis, that generates not only isoleucine and valine, but also leucine. (R) 2,3-Dihydroxy-methylvalerate is generated from 3-Hydroxy-3-methyl-2-oxopentanoic acid via the enzyme ketol-acid reductoisomerase (EC 1.1.1.86) then it is converted to (S)-3-methyl-2-oxopentanoic via the dihydroxy-acid dehydratase (EC:4.2.1.9). | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C6H12O4 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Weight: | 148.1571 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Molecular Weight: | 148.073558872 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | PDGXJDXVGMHUIR-UJURSFKZSA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C6H12O4/c1-3-6(2,10)4(7)5(8)9/h4,7,10H,3H2,1-2H3,(H,8,9)/t4-,6+/m0/s1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | 562-43-6 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | (2R,3R)-2,3-dihydroxy-3-methylpentanoic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | (2R,3R)-2,3-dihydroxy-3-methylpentanoic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | CC[C@@](C)(O)[C@@H](O)C(O)=O | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Taxonomy Description | This compound belongs to the class of organic compounds known as hydroxy fatty acids. These are fatty acids in which the chain bears a hydroxyl group. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Lipids and lipid-like molecules | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Fatty Acyls | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Fatty acids and conjugates | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Hydroxy fatty acids | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aliphatic acyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Solid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | -1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | (R) 2,3-Dihydroxy-3-methylvalerate > 3-Methyl-2-oxovaleric acid + Water 2-Aceto-2-hydroxy-butyrate + Hydrogen ion + NADPH <> (R) 2,3-Dihydroxy-3-methylvalerate + NADP (R) 2,3-Dihydroxy-3-methylvalerate + NADP <> (R)-3-Hydroxy-3-methyl-2-oxopentanoate + NADPH + Hydrogen ion (R) 2,3-Dihydroxy-3-methylvalerate + NADP > 2-Aceto-2-hydroxy-butyrate + NADPH (R)-2,3-Dihydroxy-isovalerate + Hydrogen ion + NADPH + NADPH > NADP + (R) 2,3-Dihydroxy-3-methylvalerate 2-Aceto-2-hydroxy-butyrate + NADPH + Hydrogen ion + NADPH > NADP + (R) 2,3-Dihydroxy-3-methylvalerate (R) 2,3-Dihydroxy-3-methylvalerate > Water + 3-Methyl-2-oxovaleric acid + 3-Methyl-2-oxovaleric acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||