|
Record Information |
|---|
| Version |
1.0 |
|---|
| Update Date |
1/22/2018 11:54:54 AM |
|---|
|
Metabolite ID | PAMDB000805 |
|---|
|
Identification |
|---|
| Name: |
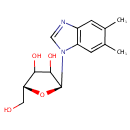
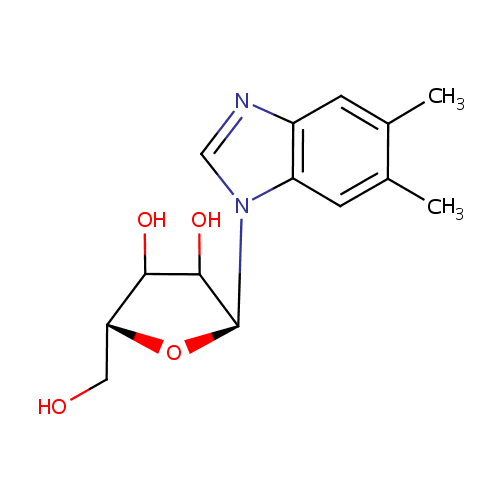
N1-(alpha-D-ribosyl)-5,6-dimethyl-benzimidazole |
|---|
| Description: | N1-(alpha-D-ribosyl)-5,6-dimethyl-benzimidazole is also known as alpha-ribazole. It is converted from N1-(5-Phospho-alpha-D-ribosyl)-5,6-dimethylbenzimidazole via dephosphorylation by the enzyme alpha-ribazole phosphatase (EC 3.1.3.73). It can subsequently be used to synthesize Vitamin B12 coenzyme. (KEGG) |
|---|
|
Structure |
|
|---|
| Synonyms: | - (2S,5R)-2-(5,6 dimethylbenzimidazol-1-yl)-5 (hydroxymethyl)oxolane-3,4-diol
- 5,6-Dimethyl-1-a-D-ribofuranosyl-1H-benzimidazole
- 5,6-Dimethyl-1-a-delta-ribofuranosyl-1H-benzimidazole
- 5,6-Dimethyl-1-a-δ-ribofuranosyl-1H-benzimidazole
- 5,6-Dimethyl-1-alpha-delta-ribofuranosyl-1H-benzimidazole
- 5,6-Dimethyl-1-α-δ-ribofuranosyl-1H-benzimidazole
- A-Ribazole
- Alpha-Ribazole
- N1-(α-D-ribosyl)-5,6-dimethylbenzimidazole
- N1-(a-D-Ribosyl)-5,6-dimethyl-benzimidazole
- N1-(a-D-Ribosyl)-5,6-dimethylbenzimidazole
- N1-(a-delta-Ribosyl)-5,6-dimethylbenzimidazole
- N1-(a-δ-Ribosyl)-5,6-dimethylbenzimidazole
- N1-(alpha-D-ribosyl)-5,6-dimethylbenzimidazole
- N1-(alpha-delta-ribosyl)-5,6-dimethylbenzimidazole
- N1-(α-D-Ribosyl)-5,6-dimethyl-benzimidazole
- N1-(α-D-Ribosyl)-5,6-dimethylbenzimidazole
- N1-(α-δ-Ribosyl)-5,6-dimethylbenzimidazole
- α-Ribazole
|
|---|
|
Chemical Formula: |
C14H18N2O4 |
|---|
| Average Molecular Weight: |
278.3037 |
|---|
| Monoisotopic Molecular
Weight: |
278.126657074 |
|---|
| InChI Key: |
HLRUKOJSWOKCPP-RYSNWHEDSA-N |
|---|
| InChI: | InChI=1S/C14H18N2O4/c1-7-3-9-10(4-8(7)2)16(6-15-9)14-13(19)12(18)11(5-17)20-14/h3-4,6,11-14,17-19H,5H2,1-2H3/t11-,12?,13?,14+/m1/s1 |
|---|
| CAS
number: |
Not Available |
|---|
| IUPAC Name: | (2S,5R)-2-(5,6-dimethyl-1H-1,3-benzodiazol-1-yl)-5-(hydroxymethyl)oxolane-3,4-diol |
|---|
|
Traditional IUPAC Name: |
(2S,5R)-2-(5,6-dimethyl-1,3-benzodiazol-1-yl)-5-(hydroxymethyl)oxolane-3,4-diol |
|---|
| SMILES: | CC1=CC2=C(C=C1C)N(C=N2)[C@H]1O[C@H](CO)C(O)C1O |
|---|
|
Chemical Taxonomy |
|---|
|
Taxonomy Description | This compound belongs to the class of organic compounds known as benzimidazole ribonucleosides and ribonucleotides. These are nucleosides with a structure that consists of an imidazole moiety of benzimidazole is N-linked to a ribose (or deoxyribose). Nucleotides have a phosphate group linked to the C5 carbon of the ribose (or deoxyribose) moiety. |
|---|
|
Kingdom |
Organic compounds |
|---|
| Super Class | Nucleosides, nucleotides, and analogues |
|---|
|
Class |
Benzimidazole ribonucleosides and ribonucleotides |
|---|
| Sub Class | Not Available |
|---|
|
Direct Parent |
Benzimidazole ribonucleosides and ribonucleotides |
|---|
| Alternative Parents |
|
|---|
| Substituents |
- 1-ribofuranosylbenzimidazole
- N-glycosyl compound
- Glycosyl compound
- Benzimidazole
- Benzenoid
- N-substituted imidazole
- Monosaccharide
- Saccharide
- Heteroaromatic compound
- Oxolane
- Imidazole
- Azole
- Secondary alcohol
- 1,2-diol
- Oxacycle
- Azacycle
- Organoheterocyclic compound
- Hydrocarbon derivative
- Primary alcohol
- Organooxygen compound
- Organonitrogen compound
- Alcohol
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework |
Aromatic heteropolycyclic compounds |
|---|
| External Descriptors |
Not Available |
|---|
|
Physical Properties |
|---|
| State: |
Solid |
|---|
| Charge: | 0 |
|---|
|
Melting point: |
Not Available |
|---|
| Experimental Properties: |
|
|---|
| Predicted Properties |
|
|---|
|
Biological Properties |
|---|
| Cellular Locations: |
Cytoplasm |
|---|
| Reactions: | |
|---|
|
Pathways: |
|
|---|
|
Spectra |
|---|
| Spectra: |
|
|---|
|
References |
|---|
| References: |
- Kanehisa, M., Goto, S., Sato, Y., Furumichi, M., Tanabe, M. (2012). "KEGG for integration and interpretation of large-scale molecular data sets." Nucleic Acids Res 40:D109-D114. Pubmed: 22080510
- Keseler, I. M., Collado-Vides, J., Santos-Zavaleta, A., Peralta-Gil, M., Gama-Castro, S., Muniz-Rascado, L., Bonavides-Martinez, C., Paley, S., Krummenacker, M., Altman, T., Kaipa, P., Spaulding, A., Pacheco, J., Latendresse, M., Fulcher, C., Sarker, M., Shearer, A. G., Mackie, A., Paulsen, I., Gunsalus, R. P., Karp, P. D. (2011). "EcoCyc: a comprehensive database of Escherichia coli biology." Nucleic Acids Res 39:D583-D590. Pubmed: 21097882
- van der Werf, M. J., Overkamp, K. M., Muilwijk, B., Coulier, L., Hankemeier, T. (2007). "Microbial metabolomics: toward a platform with full metabolome coverage." Anal Biochem 370:17-25. Pubmed: 17765195
- Winder, C. L., Dunn, W. B., Schuler, S., Broadhurst, D., Jarvis, R., Stephens, G. M., Goodacre, R. (2008). "Global metabolic profiling of Escherichia coli cultures: an evaluation of methods for quenching and extraction of intracellular metabolites." Anal Chem 80:2939-2948. Pubmed: 18331064
|
|---|
| Synthesis Reference: |
Not Available |
|---|
| Material Safety Data Sheet (MSDS) |
Not Available |
|---|
|
Links |
|---|
| External Links: |
|
|---|