| References: |
- Hazen SL, Hsu FF, d'Avignon A, Heinecke JW: Human neutrophils employ myeloperoxidase to convert alpha-amino acids to a battery of reactive aldehydes: a pathway for aldehyde generation at sites of inflammation. Biochemistry. 1998 May 12;37(19):6864-73. Pubmed: 9578573
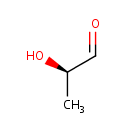
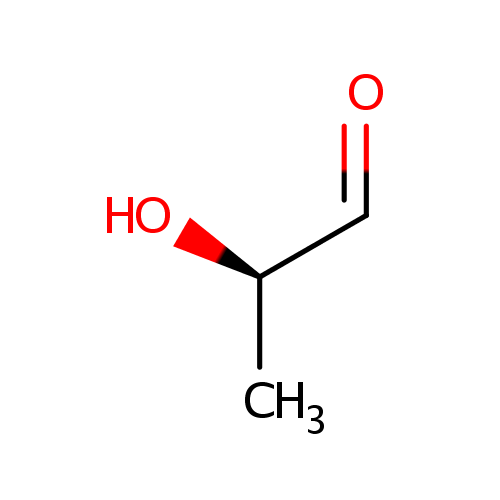
- HUANG PC, MILLER ON: The metabolism of lactaldehyde. V. Metabolism of L-fucose. J Biol Chem. 1958 Mar;231(1):201-5. Pubmed: 13538961
- Kanehisa, M., Goto, S., Sato, Y., Furumichi, M., Tanabe, M. (2012). "KEGG for integration and interpretation of large-scale molecular data sets." Nucleic Acids Res 40:D109-D114. Pubmed: 22080510
- Keseler, I. M., Collado-Vides, J., Santos-Zavaleta, A., Peralta-Gil, M., Gama-Castro, S., Muniz-Rascado, L., Bonavides-Martinez, C., Paley, S., Krummenacker, M., Altman, T., Kaipa, P., Spaulding, A., Pacheco, J., Latendresse, M., Fulcher, C., Sarker, M., Shearer, A. G., Mackie, A., Paulsen, I., Gunsalus, R. P., Karp, P. D. (2011). "EcoCyc: a comprehensive database of Escherichia coli biology." Nucleic Acids Res 39:D583-D590. Pubmed: 21097882
- Vander Jagt, D. L., Robinson, B., Taylor, K. K., Hunsaker, L. A. (1992). "Reduction of trioses by NADPH-dependent aldo-keto reductases. Aldose reductase, methylglyoxal, and diabetic complications." J Biol Chem 267:4364-4369. Pubmed: 1537826
- Winder, C. L., Dunn, W. B., Schuler, S., Broadhurst, D., Jarvis, R., Stephens, G. M., Goodacre, R. (2008). "Global metabolic profiling of Escherichia coli cultures: an evaluation of methods for quenching and extraction of intracellular metabolites." Anal Chem 80:2939-2948. Pubmed: 18331064
|
|---|