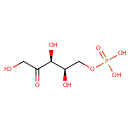
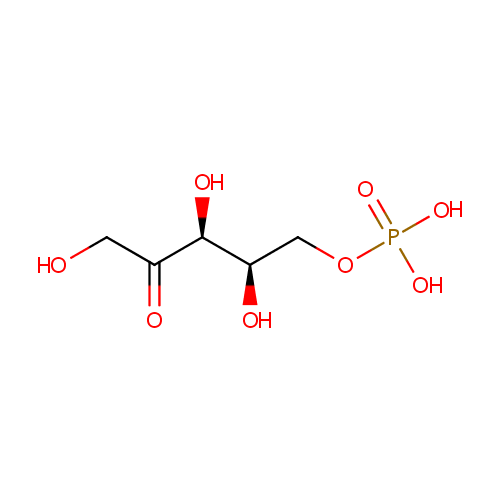
Xylulose 5-phosphate (PAMDB000653)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 1.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 1/22/2018 11:54:54 AM | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolite ID | PAMDB000653 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | Xylulose 5-phosphate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description: | Xylulose 5-phosphate (Xu-5-P) is a metabolite of the hexose monophosphate pathway that activates protein phosphatase 2A to mediate the acute effects of carbohydrate feeding on the glycolytic pathway, as well as the coordinate long-term control of the enzymes required for fatty acid and triglyceride synthesis. Xu-5-P is the signal for the coordinated control of lipogenesis. Feeding carbohydrates to cells causes levels of glucose, Glucose-6-phosphate (Glc-6-P), and Fructose-6-phosphate (Fru-6-P) to rise. Elevation of Fru-6-P leads to elevation of Xu-5-P in reactions catalyzed by the near-equilibrium isomerases of the nonoxidative portion of the hexose monophosphate pathway (ribulose 5-phosphate (Ru5P) epimerase [EC 5.1.3.1], ribose 5-phosphate (Rib5P) isomerase [EC 5.3.1.6], transaldolase [EC 2.2.1.2], and transketolase [EC 2.2.1.1]). The elevation of Xu-5-P is the coordinating signal that both acutely activates phosphofructokinase [PFK; EC 2.7.1.11] in glycolysis and will increase transcription of the genes for the enzymes of lipogenesis, the hexose monophosphate shunt, and glycolysis, all of which are required for the de novo synthesis of fat. (PMID 12721358) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C5H11O8P | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Weight: | 230.1098 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Molecular Weight: | 230.01915384 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | FNZLKVNUWIIPSJ-RFZPGFLSSA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C5H11O8P/c6-1-3(7)5(9)4(8)2-13-14(10,11)12/h4-6,8-9H,1-2H2,(H2,10,11,12)/t4-,5-/m1/s1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | 4212-65-1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | {[(2R,3S)-2,3,5-trihydroxy-4-oxopentyl]oxy}phosphonic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | ribulose-5-phosphate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | OCC(=O)[C@@H](O)[C@H](O)COP(O)(O)=O | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Taxonomy Description | This compound belongs to the class of organic compounds known as monosaccharide phosphates. These are monosaccharides comprising a phosphated group linked to the carbohydrate unit. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organooxygen compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Carbohydrates and carbohydrate conjugates | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Monosaccharides | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Monosaccharide phosphates | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aliphatic acyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Solid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | -2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | Adenosine triphosphate + D-Xylulose <> ADP + Hydrogen ion + Xylulose 5-phosphate D-Ribose-5-phosphate + Xylulose 5-phosphate <> D-Glyceraldehyde 3-phosphate + D-Sedoheptulose 7-phosphate L-Ribulose 5-phosphate <> Xylulose 5-phosphate D-Ribulose 5-phosphate <> Xylulose 5-phosphate D-Ribulose 5-phosphate <> Xylulose 5-phosphate Adenosine triphosphate + D-Xylulose <> ADP + Xylulose 5-phosphate Sedoheptulose 7-phosphate + D-Glyceraldehyde 3-phosphate + D-Sedoheptulose 7-phosphate <> D-Ribose-5-phosphate + Xylulose 5-phosphate beta-D-Fructose 6-phosphate + D-Glyceraldehyde 3-phosphate <> D-Erythrose 4-phosphate + Xylulose 5-phosphate D-Erythrose 4-phosphate + Xylulose 5-phosphate <> Fructose 6-phosphate + D-Glyceraldehyde 3-phosphate D-Ribulose 5-phosphate <> Xylulose 5-phosphate + Xylulose 5-phosphate Xylulose 5-phosphate + D-Ribose-5-phosphate + Xylulose 5-phosphate <> D-Sedoheptulose 7-phosphate + D-Glyceraldehyde 3-phosphate + D-Sedoheptulose 7-phosphate + D-Glyceraldehyde 3-phosphate L-Threo-2-pentulose + Adenosine triphosphate + L-Threo-2-pentulose > Xylulose 5-phosphate + Adenosine diphosphate + Xylulose 5-phosphate + ADP More...3-keto-L-gulonate 6-phosphate + Hydrogen ion + 3-Keto-L-gulonate 6-phosphate > Xylulose 5-phosphate + Carbon dioxide + Xylulose 5-phosphate 2,3-Diketo-L-gulonate + Hydrogen ion + 2,3-Diketo-L-gulonate > Carbon dioxide + Xylulose 5-phosphate + Xylulose 5-phosphate Xylulose 5-phosphate + Xylulose 5-phosphate > L-ribulose 5-phosphate + L-ribulose 5-phosphate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Shaeri, Jobin; Wohlgemuth, Roland; Woodley, John M. Semiquantitative Process Screening for the Biocatalytic Synthesis of D-Xylulose-5-phosphate. Organic Process Research & Development (2006), 10(3), 605-610. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Enzymes
- General function:
- Involved in catalytic activity
- Specific function:
- D-ribulose 5-phosphate = D-xylulose 5- phosphate
- Gene Name:
- rpe
- Locus Tag:
- PA0607
- Molecular weight:
- 24.1 kDa
Reactions
| D-ribulose 5-phosphate = D-xylulose 5-phosphate. |
- General function:
- Involved in catalytic activity
- Specific function:
- Sedoheptulose 7-phosphate + D-glyceraldehyde 3-phosphate = D-ribose 5-phosphate + D-xylulose 5-phosphate
- Gene Name:
- tktA
- Locus Tag:
- PA0548
- Molecular weight:
- 72.2 kDa
Reactions
| Sedoheptulose 7-phosphate + D-glyceraldehyde 3-phosphate = D-ribose 5-phosphate + D-xylulose 5-phosphate. |