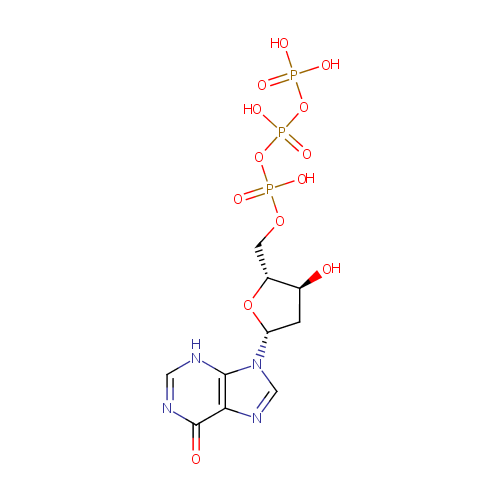
2'-Deoxyinosine triphosphate (PAMDB000593)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 1.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 1/22/2018 11:54:54 AM | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolite ID | PAMDB000593 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | 2'-Deoxyinosine triphosphate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description: | 2'-Deoxyinosine triphosphate (dITP) is a deoxyribonucleotide that may be generated from dATP by slow, non-enzymatic hydrolysis or by reduction of ITP. Normally, the cellular dITP concentration is very low. The inability to demonstrate the synthesis of dITP in cellular preparations has been attributed to the presence in the cytoplasm of an inosine triphosphatase pyrophosphatase (ITPase, EC 3.6.1.19), an enzyme that does not permit accumulation of these nucleotides. dITP can be incorporated into DNA by polymerases. The deoxyribonucleotide dITP behaves as a dGTP analogue and is incorporated opposite cytosine with about 50% efficiency. Both isolated nuclei and purified DNA polymerases rapidly incorporated dITP into DNA. In the presence of ATP, dITP is stabilized in extracts of nuclei. dITP exist in all cells and is potentially mutagenic, and the levels of these nucleotides are controlled by ITPase. The function of this ubiquitous protein family is proposed to be the elimination of minor potentially mutagenic or clastogenic purine nucleoside triphosphates from the cell. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C10H15N4O13P3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Weight: | 492.1664 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Molecular Weight: | 491.984846122 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | UFJPAQSLHAGEBL-RRKCRQDMSA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C10H15N4O13P3/c15-5-1-7(14-4-13-8-9(14)11-3-12-10(8)16)25-6(5)2-24-29(20,21)27-30(22,23)26-28(17,18)19/h3-7,15H,1-2H2,(H,20,21)(H,22,23)(H,11,12,16)(H2,17,18,19)/t5-,6+,7+/m0/s1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | 16595-02-1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | {[hydroxy({[hydroxy({[(2R,3S,5R)-3-hydroxy-5-(6-oxo-6,9-dihydro-3H-purin-9-yl)oxolan-2-yl]methoxy})phosphoryl]oxy})phosphoryl]oxy}phosphonic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | [hydroxy({hydroxy[(2R,3S,5R)-3-hydroxy-5-(6-oxo-3H-purin-9-yl)oxolan-2-yl]methoxyphosphoryl}oxy)phosphoryl]oxyphosphonic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | O[C@H]1C[C@@H](O[C@@H]1COP(O)(=O)OP(O)(=O)OP(O)(O)=O)N1C=NC2=C1NC=NC2=O | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Taxonomy Description | This compound belongs to the class of organic compounds known as purine 2'-deoxyribonucleoside triphosphates. These are purine nucleotides with triphosphate group linked to the ribose moiety lacking a hydroxyl group at position 2. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Nucleosides, nucleotides, and analogues | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Purine nucleotides | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Purine deoxyribonucleotides | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Purine 2'-deoxyribonucleoside triphosphates | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aromatic heteropolycyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Solid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | -4 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | 2'-Deoxyinosine triphosphate + Water > DIMP + Hydrogen ion + Pyrophosphate 2'-Deoxyinosine triphosphate + Water > DIDP + Hydrogen ion + Phosphate dATP + Hydrogen ion + Water > 2'-Deoxyinosine triphosphate + Ammonium Adenosine triphosphate + DIDP <> ADP + 2'-Deoxyinosine triphosphate 2'-Deoxyinosine triphosphate + Water <> DIMP + Pyrophosphate Xanthosine 5-triphosphate + Water + 2'-Deoxyinosine triphosphate <> Xanthylic acid + Pyrophosphate + DIMP | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||