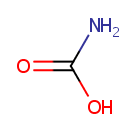
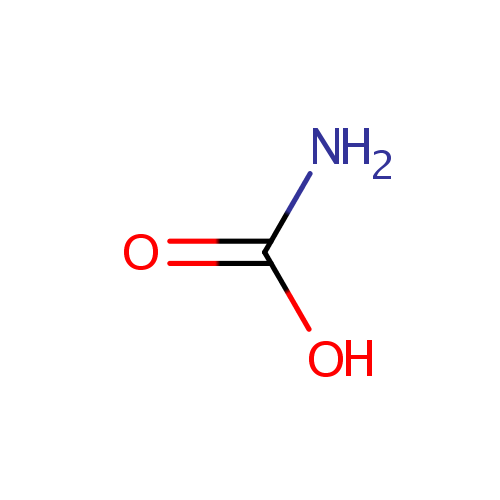
Carbamic acid (PAMDB000513)
| Record Information | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 1.0 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 1/22/2018 11:54:54 AM | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolite ID | PAMDB000513 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | Carbamic acid | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description: | Carbamic acid products of several amines, such as beta-N-methylamino-L-alanine (BMAA), ethylenediamine, and L-cysteine have been implicated in toxicity. Studies suggested that a significant portion of amino-compounds in biological samples (that naturally contain CO2/bicarbonate) can be present as a carbamic acid. There has been no obvious species specificity for their formation and no preference for 1 or 2 degree amines. Many biological reactions have also been described in the literature that involve the reaction of CO2 with amino groups of biomolecules. For example, CO2 generated from cellular respiration is expired in part through the reversible formation of a carbamate between CO2 and the -amino groups of the alpha and beta-chains of hemoglobin. The carbamic acid is also a substrate for glucuronidation and results in a stable carbamate glucuronide metabolite. The detection and characterization of these products has been facilitated greatly by the advent of soft ionization mass spectrometry techniques and high field NMR instrumentation. (PMID: 16268118, 17168688, 12929145) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | CH3NO2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Weight: | 61.04 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Molecular Weight: | 61.016378345 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | KXDHJXZQYSOELW-UHFFFAOYSA-N | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/CH3NO2/c2-1(3)4/h2H2,(H,3,4) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | 463-77-4 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | carbamic acid | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | carbamic acid | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | NC(O)=O | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Taxonomy Description | This compound belongs to the class of organic compounds known as carbamic acids. These are compounds containing the carbamic acid functional group, with the general structure RN(R')C(=O)OH or RN(R')C(=O)[O-] where R,R' = H or organyl group. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organic acids and derivatives | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Carboxylic acids and derivatives | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Amino acids, peptides, and analogues | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Carbamic acids | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aliphatic acyclic compounds | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Solid | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | -1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | Water + Ureidoacrylate peracid > Carbamic acid + Hydrogen ion + Peroxyaminoacrylate Carbamic acid + 2 Hydrogen ion > Carbon dioxide + Ammonium Adenosine triphosphate + Carbamic acid <> ADP + Carbamoylphosphate Cyanate + Hydrogen ion + Hydrogen carbonate <> Carbon dioxide + Carbamic acid Ureidoacrylate peracid + Water <> Peroxyaminoacrylate + Carbamic acid Ureidoacrylate + Water <> 3-Aminoacrylate + Carbamic acid Carbamic acid + Hydrogen ion > Ammonia + Carbon dioxide 3-ureidoacrylate + Water > Hydrogen ion + Carbamic acid + 3-Aminoacrylate (<i>Z</i>)-2-methylureidoacrylate peracid + Water > (<i>Z</i>)-2-methyl-peroxyaminoacrylate + Carbamic acid + Hydrogen ion 2 Adenosine triphosphate + L-Glutamine + Hydrogen carbonate + Water + Ammonia + Carbamic acid + Carboxyphosphate <>2 ADP + Phosphate + L-Glutamate + Carbamoylphosphate Cyanate + Hydrogen carbonate + 2 Hydrogen ion + Carbamic acid <> Ammonia +2 Carbon dioxide Ureidoacrylate peracid + Water + Carbamic acid + (Z)-2-Methyl-ureidoacrylate peracid <> Peroxyaminoacrylate + Carbon dioxide + Ammonia + (Z)-2-Methyl-peroxyaminoacrylate Hydrogen carbonate + Cyanate + Hydrogen ion + Cyanate > Carbamic acid Carbon dioxide + Water > Carbamic acid | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways: | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Download (PDF) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||