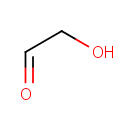
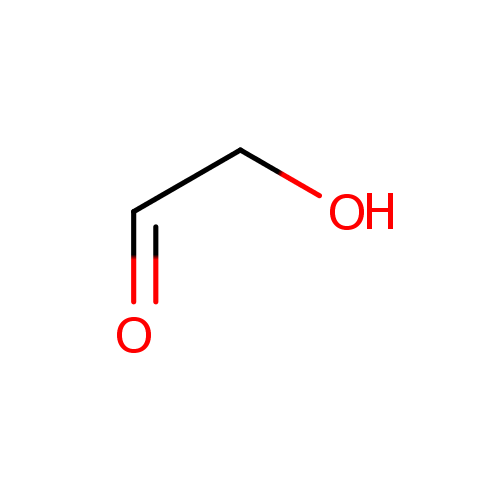
Glycolaldehyde (PAMDB000444)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 1.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 1/22/2018 11:54:54 AM | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolite ID | PAMDB000444 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | Glycolaldehyde | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description: | Glycolaldehyde (HOCH2-CH=O, IUPAC name 2-hydroxyethanal) is a type of diose (2-carbon monosaccharide). Glycolaldehyde is readily converted to acetyl coenzyme A. It has an aldehyde and a hydroxyl group. However, it is not actually a sugar, because there is only one hydroxyl group. Glycolaldehyde is formed from many sources, including the amino acid glycine and from purone catabolism. It can form by action of ketolase on fructose 1,6-bisphosphate in an alternate glycolysis pathway. This compound is transferred by thiamin pyrophosphate during the pentose phosphate shunt. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C2H4O2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Weight: | 60.052 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Molecular Weight: | 60.021129372 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | WGCNASOHLSPBMP-UHFFFAOYSA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C2H4O2/c3-1-2-4/h1,4H,2H2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | 141-46-8 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | 2-hydroxyacetaldehyde | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | glycolaldehyde | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | OCC=O | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Taxonomy Description | This compound belongs to the class of organic compounds known as short-chain aldehydes. These are an aldehyde with a chain length containing between 2 and 5 carbon atoms. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organooxygen compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Carbonyl compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Aldehydes | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Short-chain aldehydes | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aliphatic acyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Solid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | 0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | 97 °C | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | FMNH + Isethionic acid + Oxygen > Flavin Mononucleotide + Glycolaldehyde + Hydrogen ion + Water + Sulfite Glycolaldehyde + Water + NAD > Glycolic acid +2 Hydrogen ion + NADH 7,8-Dihydroneopterin <> 6-Hydroxymethyl dihydropterin + Glycolaldehyde Ethylene glycol + NAD <> Glycolaldehyde + NADH + Hydrogen ion D-Ribulose-1-phosphate <> Glycolaldehyde + Dihydroxyacetone phosphate 4-Hydroxy-L-threonine <> Glycolaldehyde + Glycine Glycolaldehyde + NAD + Water > Glycolic acid + NADH 7,8-Dihydroneopterin + 7,8-Dihydroneopterin > Glycolaldehyde + 6-Hydroxymethyl dihydropterin | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Majerski, Piotr A.; Piskorz, Jan K.; Radlein, Desmond St. A. G. Production of glycolaldehyde by hydrous thermolysis of sugars. PCT Int. Appl. (2002), 41 pp. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Download (PDF) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||