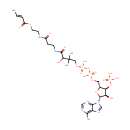
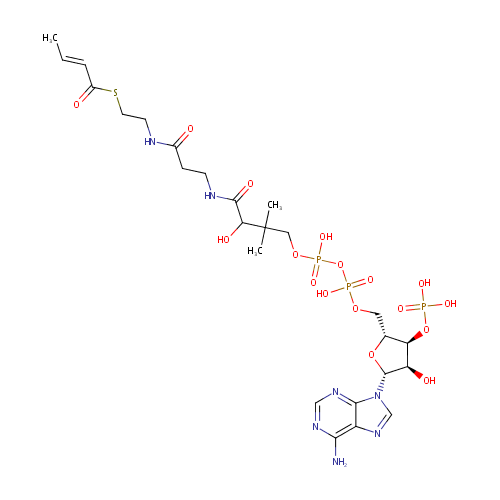
Crotonoyl-CoA (PAMDB000429)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 1.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 1/22/2018 11:54:54 AM | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolite ID | PAMDB000429 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | Crotonoyl-CoA | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description: | Crotonoyl-CoA is an important component in several metabolic pathways, notably fatty acid and amino acid metabolism. It is the substrate of a group of enzymes acyl-Coenzyme A oxidases 1, 2, 3 (E.C.: 1.3.3.6) corresponding to palmitoyl, branched chain, and pristanoyl, respectively, in the peroxisomal fatty acid beta-oxidation, producing hydrogen peroxide. It is also a substrate of a group of enzymes called acyl-Coenzyme A dehydrogenase (E.C.:1.3.99-, including 1.3.99.2, 1.3.99.3) in the metabolism of fatty acids. In addition, crotonoyl-CoA is the substrate of enoyl coenzyme A hydratase (E.C.4.2.1.17) during lysine degradation and tryptophan metabolism, benzoate degradation via CoA ligation; in contrast it is the product of this enzyme in the butanoate metabolism. Moreover, it is produced from multiple enzymes in the butanoate metabolism pathway, including 3-Hydroxybutyryl-CoA dehydratase (E.C.:4.2.1.55), glutaconyl-CoA decarboxylase (E.C.: 4.1.1.70), vinylacetyl-CoA delta-isomerase (E.C.: 5.3.3.3), and trans-2-enoyl-CoA reductase (NAD+) (E.C.: 1.3.1.44). In lysine degradation and tryptophan metabolism, crotonoyl CoA is produced by glutaryl-Coenzyme A dehydrogenase (E.C.:1.3.99.7) lysine and tryptophan metabolic pathway. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C25H40N7O17P3S | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Weight: | 835.608 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Molecular Weight: | 835.141423115 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | KFWWCMJSYSSPSK-BOGFJHSMSA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C25H40N7O17P3S/c1-4-5-16(34)53-9-8-27-15(33)6-7-28-23(37)20(36)25(2,3)11-46-52(43,44)49-51(41,42)45-10-14-19(48-50(38,39)40)18(35)24(47-14)32-13-31-17-21(26)29-12-30-22(17)32/h4-5,12-14,18-20,24,35-36H,6-11H2,1-3H3,(H,27,33)(H,28,37)(H,41,42)(H,43,44)(H2,26,29,30)(H2,38,39,40)/b5-4+/t14-,18-,19-,20?,24-/m1/s1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | 102680-35-3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | {[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-2-{[({[(3-{[2-({2-[(2E)-but-2-enoylsulfanyl]ethyl}carbamoyl)ethyl]carbamoyl}-3-hydroxy-2,2-dimethylpropoxy)(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl)oxy]methyl}-4-hydroxyoxolan-3-yl]oxy}phosphonic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | [(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-2-({[(3-{[2-({2-[(2E)-but-2-enoylsulfanyl]ethyl}carbamoyl)ethyl]carbamoyl}-3-hydroxy-2,2-dimethylpropoxy(hydroxy)phosphoryl)oxy(hydroxy)phosphoryl]oxy}methyl)-4-hydroxyoxolan-3-yl]oxyphosphonic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | C\C=C\C(=O)SCCNC(=O)CCNC(=O)C(O)C(C)(C)COP(O)(=O)OP(O)(=O)OC[C@H]1O[C@H]([C@H](O)[C@@H]1OP(O)(O)=O)N1C=NC2=C(N)N=CN=C12 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Taxonomy Description | This compound belongs to the class of organic compounds known as acyl coas. These are organic compounds containing a coenzyme A substructure linked to an acyl chain. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Lipids and lipid-like molecules | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Fatty Acyls | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Fatty acyl thioesters | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Acyl CoAs | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aromatic heteropolycyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Solid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | -4 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | 3-Hydroxybutyryl-CoA <> Crotonoyl-CoA + Water Butyryl-CoA + FAD <> Crotonoyl-CoA + FADH2 Butanoyl-CoA + FAD <> FADH2 + Crotonoyl-CoA (S)-3-Hydroxybutanoyl-CoA <> Crotonoyl-CoA + Water Crotonoyl-CoA + Water <> 3-Hydroxybutanoyl-CoA Crotonoyl-CoA + Water > (3S)-3-hydroxyacyl-CoA + (3S)-3-Hydroxyacyl-CoA Crotonoyl-CoA > Water + 3-Hydroxybutyryl-CoA + 3-Hydroxybutyryl-CoA Butyryl-CoA + electron-transfer flavoprotein + Butyryl-CoA <> Crotonoyl-CoA + Reduced electron-transfer flavoprotein Glutaryl-CoA > Crotonoyl-CoA + Carbon dioxide 3-Hydroxybutyryl-CoA + 3-Hydroxybutyryl-CoA > Crotonoyl-CoA | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||