|
Record Information |
|---|
| Version |
1.0 |
|---|
| Update Date |
1/22/2018 11:54:54 AM |
|---|
|
Metabolite ID | PAMDB000366 |
|---|
|
Identification |
|---|
| Name: |
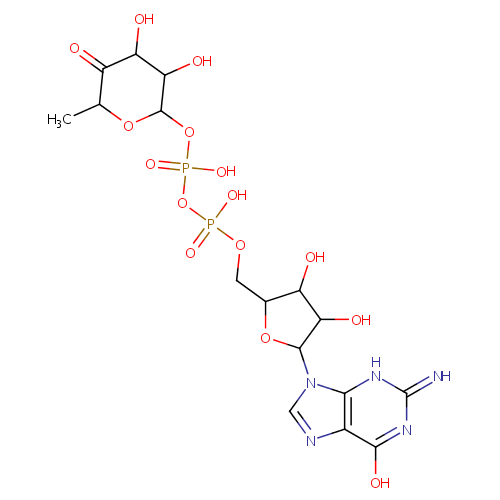
GDP-4-Dehydro-6-L-deoxygalactose |
|---|
| Description: | GDP-4-dehydro-6-L-deoxygalactose is a member of the chemical class known as Purine Nucleotide Sugars. These are purine nucleotides bound to a saccharide derivative through the terminal phosphate group. GDP-6-deoxy-D-talose biosynthetic pathway involve the enzyme GDP-4-keto-6-deoxy-D-mannose-3-dehydratase, which catalyzes the third step in colitose production, which is the removal of the hydroxyl group at C3' of GDP-4-keto-6-deoxymannose. (PMID 16943443) |
|---|
|
Structure |
|
|---|
| Synonyms: | - GDP-4-Dehydro-6-deoxy-D-talose
- GDP-4-Dehydro-6-L-deoxygalactose
- GDP-4-Keto-6-deoxy-D-mannose
- GDP-4-Keto-6-deoxymannose
- GDP-4-Keto-6-L-deoxygalactose
- GDP-4-Oxo-6-deoxy-D-mannose
- GDP-4-Oxo-6-deoxymannose
- GDP-DDMan
|
|---|
|
Chemical Formula: |
C16H23N5O15P2 |
|---|
| Average Molecular Weight: |
587.3258 |
|---|
| Monoisotopic Molecular
Weight: |
587.066588115 |
|---|
| InChI Key: |
PNHLMHWWFOPQLK-GTXIGXBOSA-N |
|---|
| InChI: | InChI=1S/C16H23N5O15P2/c1-4-7(22)9(24)11(26)15(33-4)35-38(30,31)36-37(28,29)32-2-5-8(23)10(25)14(34-5)21-3-18-6-12(21)19-16(17)20-13(6)27/h3-5,8-11,14-15,23-26H,2H2,1H3,(H,28,29)(H,30,31)(H3,17,19,20,27)/t4-,5?,8+,9+,10+,11-,14+,15?/m0/s1 |
|---|
| CAS
number: |
Not Available |
|---|
| IUPAC Name: | [({[3,4-dihydroxy-5-(6-hydroxy-2-imino-3,9-dihydro-2H-purin-9-yl)oxolan-2-yl]methoxy}(hydroxy)phosphoryl)oxy][(3,4-dihydroxy-6-methyl-5-oxooxan-2-yl)oxy]phosphinic acid |
|---|
|
Traditional IUPAC Name: |
{[3,4-dihydroxy-5-(6-hydroxy-2-imino-3H-purin-9-yl)oxolan-2-yl]methoxy(hydroxy)phosphoryl}oxy(3,4-dihydroxy-6-methyl-5-oxooxan-2-yl)oxyphosphinic acid |
|---|
| SMILES: | C[C@@H]1OC(OP(O)(=O)OP(O)(=O)OCC2O[C@H]([C@H](O)[C@@H]2O)N2C=NC3=C2N=C(N)NC3=O)[C@@H](O)[C@H](O)C1=O |
|---|
|
Chemical Taxonomy |
|---|
|
Taxonomy Description | This compound belongs to the class of organic compounds known as purine nucleotide sugars. These are purine nucleotides bound to a saccharide derivative through the terminal phosphate group. |
|---|
|
Kingdom |
Organic compounds |
|---|
| Super Class | Nucleosides, nucleotides, and analogues |
|---|
|
Class |
Purine nucleotides |
|---|
| Sub Class | Purine nucleotide sugars |
|---|
|
Direct Parent |
Purine nucleotide sugars |
|---|
| Alternative Parents |
|
|---|
| Substituents |
- Purine nucleotide sugar
- Purine ribonucleoside diphosphate
- N-glycosyl compound
- Glycosyl compound
- Organic pyrophosphate
- Monosaccharide phosphate
- Purine
- Imidazopyrimidine
- Hydroxypyrimidine
- Monoalkyl phosphate
- Alkyl phosphate
- Pyrimidine
- Phosphoric acid ester
- Oxane
- Organic phosphoric acid derivative
- Organic phosphate
- N-substituted imidazole
- Monosaccharide
- Saccharide
- Heteroaromatic compound
- Oxolane
- Imidazole
- Azole
- Cyclic ketone
- Secondary alcohol
- Ketone
- 1,2-diol
- Oxacycle
- Azacycle
- Organoheterocyclic compound
- Hydrocarbon derivative
- Organooxygen compound
- Organonitrogen compound
- Carbonyl group
- Alcohol
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework |
Aromatic heteropolycyclic compounds |
|---|
| External Descriptors |
Not Available |
|---|
|
Physical Properties |
|---|
| State: |
Not Available |
|---|
| Charge: | -2 |
|---|
|
Melting point: |
Not Available |
|---|
| Experimental Properties: |
|
|---|
| Predicted Properties |
|
|---|
|
Biological Properties |
|---|
| Cellular Locations: |
Cytoplasm |
|---|
| Reactions: | |
|---|
|
Pathways: |
- Amino sugar and nucleotide sugar metabolism pae00520
- Fructose and mannose metabolism pae00051
|
|---|
|
Spectra |
|---|
| Spectra: |
|
|---|
|
References |
|---|
| References: |
- Cook, P. D., Thoden, J. B., Holden, H. M. (2006). "The structure of GDP-4-keto-6-deoxy-D-mannose-3-dehydratase: a unique coenzyme B6-dependent enzyme." Protein Sci 15:2093-2106. Pubmed: 16943443
- Kanehisa, M., Goto, S., Sato, Y., Furumichi, M., Tanabe, M. (2012). "KEGG for integration and interpretation of large-scale molecular data sets." Nucleic Acids Res 40:D109-D114. Pubmed: 22080510
- Keseler, I. M., Collado-Vides, J., Santos-Zavaleta, A., Peralta-Gil, M., Gama-Castro, S., Muniz-Rascado, L., Bonavides-Martinez, C., Paley, S., Krummenacker, M., Altman, T., Kaipa, P., Spaulding, A., Pacheco, J., Latendresse, M., Fulcher, C., Sarker, M., Shearer, A. G., Mackie, A., Paulsen, I., Gunsalus, R. P., Karp, P. D. (2011). "EcoCyc: a comprehensive database of Escherichia coli biology." Nucleic Acids Res 39:D583-D590. Pubmed: 21097882
- van der Werf, M. J., Overkamp, K. M., Muilwijk, B., Coulier, L., Hankemeier, T. (2007). "Microbial metabolomics: toward a platform with full metabolome coverage." Anal Biochem 370:17-25. Pubmed: 17765195
|
|---|
| Synthesis Reference: |
Not Available |
|---|
| Material Safety Data Sheet (MSDS) |
Not Available |
|---|
|
Links |
|---|
| External Links: |
|
|---|