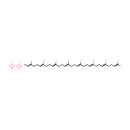Octaprenyl diphosphate (PAMDB000251)
| Record Information | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 1.0 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 1/22/2018 11:54:54 AM | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolite ID | PAMDB000251 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | Octaprenyl diphosphate | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description: | Octaprenyl diphosphate belongs to the class of Tetraterpenes. These are terpene molecules containing 10 consecutively linked isoprene units. (inferred from compound structure)Octaprenyl diphosphate is invovled in Terpenoid backbone biosynthesis, Ubiquinone and other terpenoid-quinone biosynthesis, and Biosynthesis of secondary metabolites. (KEGG) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C40H68O7P2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Weight: | 722.9112 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Molecular Weight: | 722.444027554 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | IKKLDISSULFFQO-DJMILUHSSA-N | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C40H68O7P2/c1-33(2)17-10-18-34(3)19-11-20-35(4)21-12-22-36(5)23-13-24-37(6)25-14-26-38(7)27-15-28-39(8)29-16-30-40(9)31-32-46-49(44,45)47-48(41,42)43/h17,19,21,23,25,27,29,31H,10-16,18,20,22,24,26,28,30,32H2,1-9H3,(H,44,45)(H2,41,42,43)/b34-19+,35-21+,36-23+,37-25+,38-27+,39-29+,40-31+ | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | {[hydroxy({[(2E,6E,10E,14E,18E,22E,26E)-3,7,11,15,19,23,27,31-octamethyldotriaconta-2,6,10,14,18,22,26,30-octaen-1-yl]oxy})phosphoryl]oxy}phosphonic acid | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | octaprenyl pyrophosphate | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | CC(C)=CCC\C(C)=C\CC\C(C)=C\CC\C(C)=C\CC\C(C)=C\CC\C(C)=C\CC\C(C)=C\CC\C(C)=C\COP(O)(=O)OP(O)(O)=O | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Taxonomy Description | This compound belongs to the class of organic compounds known as bactoprenol diphosphates. These are polyprenyl compounds consisting of a diphosphate group substituted by a bactoprenyl moiety. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Lipids and lipid-like molecules | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Prenol lipids | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Polyprenols | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Bactoprenol diphosphates | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aliphatic acyclic compounds | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | -2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Membrane | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | 1,4-Dihydroxy-2-naphthoic acid + Hydrogen ion + Octaprenyl diphosphate > 2-Demethylmenaquinol 8 + Carbon dioxide + Pyrophosphate 4-Hydroxybenzoic acid + Octaprenyl diphosphate > 3-Octaprenyl-4-hydroxybenzoate + Pyrophosphate 1,4-Dihydroxy-2-naphthoic acid + Octaprenyl diphosphate <> 2-Demethylmenaquinone 8 + Pyrophosphate + Carbon dioxide Farnesyl pyrophosphate + 5 Isopentenyl pyrophosphate <> Octaprenyl diphosphate +5 Pyrophosphate 4-Hydroxybenzoic acid + Octaprenyl diphosphate > 3-Octaprenyl-4-hydroxybenzoate Farnesyl pyrophosphate + 5 Isopentenyl pyrophosphate + Farnesyl pyrophosphate + 5 Isopentenyl pyrophosphate >5 Pyrophosphate + Octaprenyl diphosphate + Octaprenyl diphosphate 4-Hydroxybenzoic acid + Octaprenyl diphosphate + Octaprenyl diphosphate > Pyrophosphate + 3-Octaprenyl-4-hydroxybenzoate Octaprenyl diphosphate + 1,4-dihydroxy-2-naphthoate + Hydrogen ion + Octaprenyl diphosphate + 1,4-Dihydroxy-2-naphthoyl-CoA > Carbon dioxide + Pyrophosphate + 2-Demethylmenaquinol 8 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways: | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||