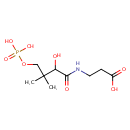
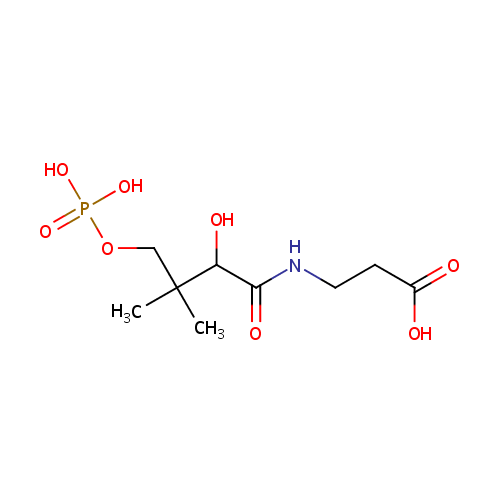
D-4'-Phosphopantothenate (PAMDB000227)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 1.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 1/22/2018 11:54:54 AM | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolite ID | PAMDB000227 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | D-4'-Phosphopantothenate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description: | D-4'-Phosphopantothenate is a product of the phosphorylatation of Pantothenate by Pantothenate kinase. It is an intermediate in coenzyme A (CoA) biosynthesis pathway. Coenzyme A is a cofactor of ubiquitous occurrence in plants, bacteria, and animals. It is needed in a large number of enzymatic reactions central to intermediary metabolism, including the oxidation of fatty acids, carbohydrates, and amino acids. Only plants and microorganisms like Pseudomonas aeruginosa are capable of synthesizing this compound de novo. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C9H18NO8P | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Weight: | 299.2149 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Molecular Weight: | 299.077003069 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | XHFVGHPGDLDEQO-UHFFFAOYSA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C9H18NO8P/c1-9(2,5-18-19(15,16)17)7(13)8(14)10-4-3-6(11)12/h7,13H,3-5H2,1-2H3,(H,10,14)(H,11,12)(H2,15,16,17) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | 3-{2-hydroxy-3-methyl-3-[(phosphonooxy)methyl]butanamido}propanoic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | 3-{2-hydroxy-3-methyl-3-[(phosphonooxy)methyl]butanamido}propanoic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | CC(C)(COP(O)(O)=O)C(O)C(=O)NCCC(O)=O | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Taxonomy Description | This compound belongs to the class of organic compounds known as beta amino acids and derivatives. These are amino acids having a (-NH2) group attached to the beta carbon atom. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organic acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Carboxylic acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Amino acids, peptides, and analogues | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Beta amino acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aliphatic acyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Solid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | -3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | D-4'-Phosphopantothenate + Cytidine triphosphate + L-Cysteine > 4-Phosphopantothenoylcysteine + Cytidine monophosphate + Hydrogen ion + Pyrophosphate Adenosine triphosphate + Pantothenic acid <> D-4'-Phosphopantothenate + ADP + Hydrogen ion Adenosine triphosphate + Pantothenic acid <> ADP + D-4'-Phosphopantothenate Adenosine triphosphate + D-4'-Phosphopantothenate + L-Cysteine <> Adenosine monophosphate + Pyrophosphate + 4-Phosphopantothenoylcysteine Cytidine triphosphate + D-4'-Phosphopantothenate + L-Cysteine <> Cytidine monophosphate + Pyrophosphate + 4-Phosphopantothenoylcysteine Pantothenic acid + Adenosine triphosphate + Pantothenic acid > D-4'-Phosphopantothenate + Adenosine diphosphate + D-4'-Phosphopantothenate + ADP Cytidine triphosphate + D-4'-Phosphopantothenate + L-Cysteine + D-4'-Phosphopantothenate > Cytidine monophosphate + Pyrophosphate + 4-Phosphopantothenoylcysteine + Cytidine monophosphate Pantothenic acid + Adenosine triphosphate + Pantothenic acid > Adenosine diphosphate + Hydrogen ion + D-4'-Phosphopantothenate + ADP + D-4'-Phosphopantothenate D-4'-Phosphopantothenate + Cytidine triphosphate + L-Cysteine + D-4'-Phosphopantothenate > Cytidine monophosphate + Pyrophosphate + Hydrogen ion + 4-Phosphopantothenoylcysteine + Cytidine monophosphate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||