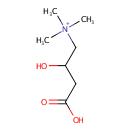
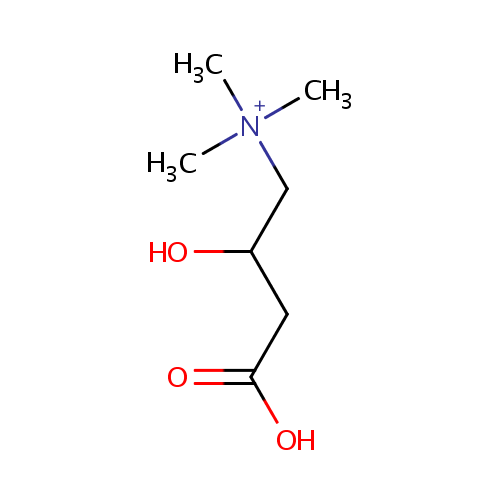
Carnitine (PAMDB000023)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 1.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 1/22/2018 11:54:54 AM | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolite ID | PAMDB000023 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | Carnitine | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description: | Carnitine is a quaternary ammonium compound biosynthesized from the amino acids lysine and methionine. In living cells, it is required for the transport of fatty acids during the breakdown of lipids (fats) for the generation of metabolic energy. Carnitine was originally found as a growth factor for mealworms and labeled vitamin Bt. Carnitine exists in two stereoisomers: Its biologically active form is L-carnitine, whereas its enantiomer, D-carnitine, is biologically inactive. Pseudomonas aeruginosa converts carnitine, via crotonobetaine, to gamma-butyrobetaine in the presence of C and N sources and under anaerobic conditions. This two-step pathway requires L-(-)-carnitine dehydratase and crotonobetaine reductase. Anaerobic carnitine metabolism in Pseudomonas aeruginosa involves six genes organized in the cai operon and located at the first minute on the Pseudomonas aeruginosa chromosome. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C7H16NO3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Weight: | 162.2068 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Molecular Weight: | 162.113018383 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | PHIQHXFUZVPYII-UHFFFAOYSA-O | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C7H15NO3/c1-8(2,3)5-6(9)4-7(10)11/h6,9H,4-5H2,1-3H3/p+1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | 541-15-1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | (3-carboxy-2-hydroxypropyl)trimethylazanium | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | (+-)-carnitine | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | C[N+](C)(C)CC(O)CC(O)=O | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Taxonomy Description | This compound belongs to the class of organic compounds known as hydroxy fatty acids. These are fatty acids in which the chain bears a hydroxyl group. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Lipids and lipid-like molecules | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Fatty Acyls | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Fatty acids and conjugates | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Hydroxy fatty acids | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aliphatic acyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Solid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | 0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | 210-212 °C | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | Adenosine triphosphate + Water + Carnitine > ADP + Carnitine + Hydrogen ion + Phosphate Adenosine triphosphate + Water + Carnitine > ADP + Carnitine + Hydrogen ion + Phosphate -->-->Adenosine triphosphate + Coenzyme A + Carnitine > ADP + L-Carnitinyl-CoA + Phosphate Adenosine triphosphate + Coenzyme A + Carnitine > ADP + D-Carnitinyl-CoA + Phosphate Carnitine + Crotonobetainyl-CoA <> L-Carnitinyl-CoA + Crotonobetaine gamma-Butyrobetainyl-CoA + Carnitine <> L-Carnitinyl-CoA + gamma-Butyrobetaine Coenzyme A + Carnitine + Adenosine triphosphate > L-Carnitinyl-CoA + Adenosine monophosphate + Pyrophosphate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Download (PDF) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Enzymes
- General function:
- Involved in nucleotide binding
- Specific function:
- Involved in a multicomponent binding-protein-dependent transport system for glycine betaine/L-proline
- Gene Name:
- proV
- Locus Tag:
- PA5094
- Molecular weight:
- 30.7 kDa